7Z0X
 
 | | THSC20.HVTR26 Fab bound to SARS-CoV-2 Receptor Binding Domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Spike protein S1, ... | | Authors: | Wibmer, C.K. | | Deposit date: | 2022-02-23 | | Release date: | 2022-04-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A combination of potently neutralizing monoclonal antibodies isolated from an Indian convalescent donor protects against the SARS-CoV-2 Delta variant.
Plos Pathog., 18, 2022
|
|
8D73
 
 | | Crystal Structure of EGFR LRTM with compound 7 | | Descriptor: | (3S,4R)-3-fluoro-1-(4-{[4-(methylamino)-1-(propan-2-yl)pyrido[3,4-d]pyridazin-7-yl]amino}pyrimidin-2-yl)piperidin-4-ol, Epidermal growth factor receptor, GLYCEROL | | Authors: | Kim, J.L. | | Deposit date: | 2022-06-07 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Discovery of BLU-945, a Reversible, Potent, and Wild-Type-Sparing Next-Generation EGFR Mutant Inhibitor for Treatment-Resistant Non-Small-Cell Lung Cancer.
J.Med.Chem., 65, 2022
|
|
8D76
 
 | | Crystal Structure of EGFR LRTM with compound 24 | | Descriptor: | (3S,4R)-3-fluoro-1-(4-{[8-{3-[(methanesulfonyl)methyl]azetidin-1-yl}-5-(propan-2-yl)-2,7-naphthyridin-3-yl]amino}pyrimidin-2-yl)-3-methylpiperidin-4-ol, Epidermal growth factor receptor, GLYCEROL | | Authors: | Kim, J.L. | | Deposit date: | 2022-06-07 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of BLU-945, a Reversible, Potent, and Wild-Type-Sparing Next-Generation EGFR Mutant Inhibitor for Treatment-Resistant Non-Small-Cell Lung Cancer.
J.Med.Chem., 65, 2022
|
|
7WVL
 
 | |
8QBV
 
 | |
8QBR
 
 | |
8CAF
 
 | | N8C_Fab3b in complex with NEDD8-CUL1(WHB) | | Descriptor: | Cullin-1, Fab Heavy Chain, Fab Light Chain, ... | | Authors: | Duda, D.M, Yanishevski, D, Henneberg, L.T, Schulman, B.A. | | Deposit date: | 2023-01-24 | | Release date: | 2023-09-13 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Activity-based profiling of cullin-RING E3 networks by conformation-specific probes.
Nat.Chem.Biol., 19, 2023
|
|
7ROZ
 
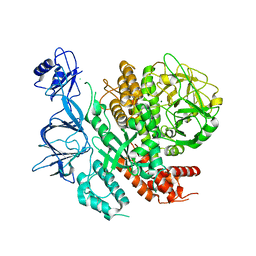 | |
7RQS
 
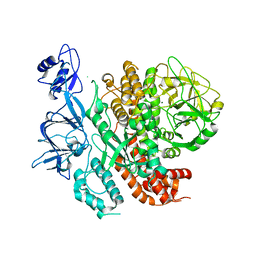 | |
3KN7
 
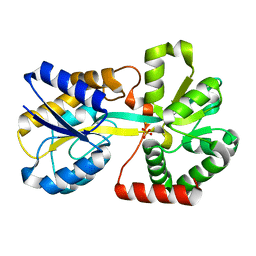 | |
3KN8
 
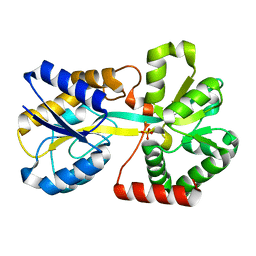 | |
1BAK
 
 | |
3FH7
 
 | |
3FH5
 
 | |
3FH8
 
 | |
3FHE
 
 | |
3FUL
 
 | |
3FTZ
 
 | | Leukotriene A4 hydrolase in complex with fragment 2-(pyridin-3-ylmethoxy)aniline | | Descriptor: | 2-(pyridin-3-ylmethoxy)aniline, ACETATE ION, IMIDAZOLE, ... | | Authors: | Davies, D.R. | | Deposit date: | 2009-01-13 | | Release date: | 2010-01-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of 4-[(2S)-2-{[4-(4-chlorophenoxy)phenoxy]methyl}-1-pyrrolidinyl]butanoic acid (DG-051) as a novel leukotriene A4 hydrolase inhibitor of leukotriene B4 biosynthesis.
J.Med.Chem., 53, 2010
|
|
3FTS
 
 | | Leukotriene A4 hydrolase in complex with resveratrol | | Descriptor: | ACETATE ION, IMIDAZOLE, Leukotriene A-4 hydrolase, ... | | Authors: | Davies, D.R. | | Deposit date: | 2009-01-13 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Discovery of leukotriene A4 hydrolase inhibitors using metabolomics biased fragment crystallography.
J.Med.Chem., 52, 2009
|
|
3FU6
 
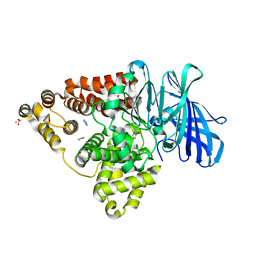 | |
3FUN
 
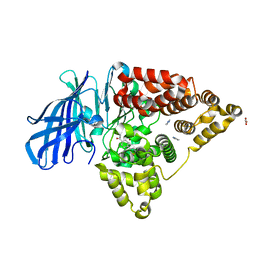 | |
3FTW
 
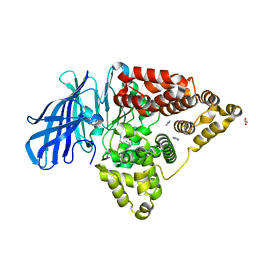 | |
3FUD
 
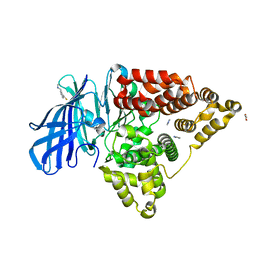 | |
3FTY
 
 | |
2QYG
 
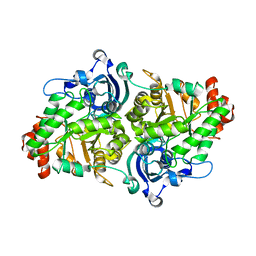 | | Crystal Structure of a RuBisCO-like Protein rlp2 from Rhodopseudomonas palustris | | Descriptor: | Ribulose bisphosphate carboxylase-like protein 2 | | Authors: | Li, H, Chan, S, Tabita, F.R, Eisenberg, D. | | Deposit date: | 2007-08-14 | | Release date: | 2007-09-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Function, structure, and evolution of the RubisCO-like proteins and their RubisCO homologs.
Microbiol.Mol.Biol.Rev., 71, 2007
|
|
