2NXV
 
 | |
2QGI
 
 | |
7P1Y
 
 | |
4UR9
 
 | | Structure of ligand bound glycosylhydrolase | | Descriptor: | 4-ethoxyquinazoline, CALCIUM ION, O-(2-ACETAMIDO-2-DEOXY D-GLUCOPYRANOSYLIDENE) AMINO-N-PHENYLCARBAMATE, ... | | Authors: | Darby, J.F, Landstroem, J, Roth, C, He, Y, Schultz, M, Davies, G.J, Hubbard, R.E. | | Deposit date: | 2014-06-27 | | Release date: | 2015-02-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of Selective Small-Molecule Activators of a Bacterial Glycoside Hydrolase.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
6TVQ
 
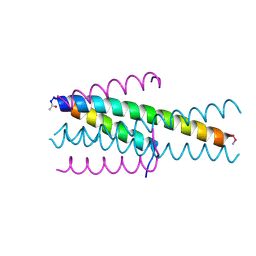 | | Structure of native gp41 derived peptide fusion inhibitor | | Descriptor: | Env polyprotein (Fragment), Envelope glycoprotein gp160 | | Authors: | Huhmann, S, Nyakatura, E.K, Rohrhofer, A, Schmidt, B, Eichler, J, Moschner, J, Roth, C. | | Deposit date: | 2020-01-10 | | Release date: | 2021-01-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Systematic Evaluation of Fluorination as Modification for Peptide-Based Fusion Inhibitors against HIV-1 Infection.
Chembiochem, 22, 2021
|
|
6TVU
 
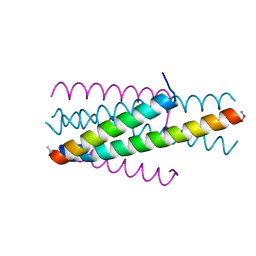 | | Structure of native gp41 derived peptide fusion inhibitor | | Descriptor: | Env polyprotein (Fragment), Transmembrane protein gp41 | | Authors: | Huhmann, S, Nyakatura, E.K, Rohrhofer, A, Schmidt, B, Eichler, J, Moschner, J, Roth, C, Koksch, B. | | Deposit date: | 2020-01-10 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Systematic Evaluation of Fluorination as Modification for Peptide-Based Fusion Inhibitors against HIV-1 Infection.
Chembiochem, 22, 2021
|
|
6TVW
 
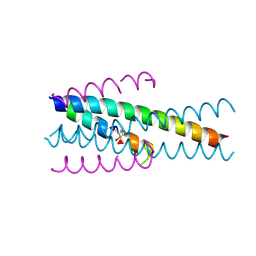 | | Structure of native gp41 derived peptide fusion inhibitor | | Descriptor: | Envelope glycoprotein, Transmembrane protein gp41,Envelope glycoprotein gp160 | | Authors: | Huhmann, S, Nyakatura, E.K, Rohrhofer, A, Schmidt, B, Eichler, J, Moschner, J, Roth, C, Koksch, B. | | Deposit date: | 2020-01-10 | | Release date: | 2021-01-27 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Systematic Evaluation of Fluorination as Modification for Peptide-Based Fusion Inhibitors against HIV-1 Infection.
Chembiochem, 22, 2021
|
|
5FL1
 
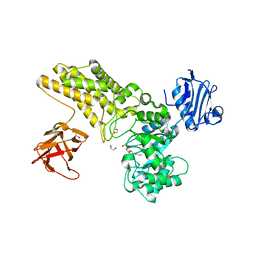 | | Structure of a hydrolase with an inhibitor | | Descriptor: | (3~{a}~{R},5~{R},6~{S},7~{R},7~{a}~{R})-5-(hydroxymethyl)-2-(prop-2-enylamino)-5,6,7,7~{a}-tetrahydro-3~{a}~{H}-pyrano[3,2-d][1,3]thiazole-6,7-diol, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Cekic, N, Heinonen, J.E, Stubbs, K.A, Roth, C, McEachern, E.J, Davies, G.J, Vocadlo, D.J. | | Deposit date: | 2015-10-20 | | Release date: | 2016-08-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Analysis of transition state mimicry by tight binding aminothiazoline inhibitors provides insight into catalysis by humanO-GlcNAcase.
Chem Sci, 7, 2016
|
|
5FL0
 
 | | Structure of a hydrolase with an inhibitor | | Descriptor: | (3~{a}~{R},5~{R},6~{S},7~{R},7~{a}~{R})-2-(butylamino)-5-(hydroxymethyl)-5,6,7,7~{a}-tetrahydro-3~{a}~{H}-pyrano[3,2-d] [1,3]thiazole-6,7-diol, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Cekic, N, Heinonen, J.E, Stubbs, K.A, Roth, C, McEachern, E.J, Davies, G.J, Vocadlo, D.J. | | Deposit date: | 2015-10-20 | | Release date: | 2016-08-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Analysis of transition state mimicry by tight binding aminothiazoline inhibitors provides insight into catalysis by humanO-GlcNAcase.
Chem Sci, 7, 2016
|
|
5FKY
 
 | | Structure of a hydrolase bound with an inhibitor | | Descriptor: | (3aR,5R,6S,7R,7aR)-2-amino-5-(hydroxymethyl)-5,6,7,7a-tetrahydro-3aH-pyrano[3,2-d][1,3]thiazole-6,7-diol, GLYCEROL, O-GLCNACASE BT_4395 | | Authors: | Cekic, N, Heinonen, J.E, Stubbs, K.A, Roth, C, McEachern, E.J, Davies, G.J, Vocadlo, D.J. | | Deposit date: | 2015-10-20 | | Release date: | 2016-01-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Analysis of transition state mimicry by tight binding aminothiazoline inhibitors provides insight into catalysis by humanO-GlcNAcase.
Chem Sci, 7, 2016
|
|
5ABH
 
 | | Structure of GH84 with ligand | | Descriptor: | 1,2-ETHANEDIOL, 2-[(2R,3S,4R,5R)-5-(hydroxymethyl)-3,4-bis(oxidanyl)-1-pentyl-pyrrolidin-2-yl]-N-methyl-ethanamide, CALCIUM ION, ... | | Authors: | Bergeron-Brlek, M, Goodwin-Tindall, J, Cekic, N, Varghese, V, Zandberg, W.F, Shan, X, Roth, C, Chan, S, Davies, G.J, Vocadlo, D.J, Britton, R. | | Deposit date: | 2015-08-05 | | Release date: | 2015-11-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Convenient Approach to Stereoisomeric Iminocyclitols: Generation of Potent Brain-Permeable Oga Inhibitors.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5ABG
 
 | | Structure of GH84 with ligand | | Descriptor: | 1,2-ETHANEDIOL, 2-[(2R,3S,4R,5R)-1-[3-(4-fluorophenyl)propyl]-5-(hydroxymethyl)-3,4-bis(oxidanyl)pyrrolidin-2-yl]-N-methyl-ethanamide, CALCIUM ION, ... | | Authors: | Bergeron-Brlek, M, Goodwin-Tindall, J, Cekic, N, Varghese, V, Zandberg, W.F, Shan, X, Roth, C, Chan, S, Davies, G.J, Vocadlo, D.J, Britton, R. | | Deposit date: | 2015-08-05 | | Release date: | 2015-11-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Convenient Approach to Stereoisomeric Iminocyclitols: Generation of Potent Brain-Permeable Oga Inhibitors.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5ABF
 
 | | Structure of GH84 with ligand | | Descriptor: | 1,2-ETHANEDIOL, 2-[(2S,3R,4R,5R)-5-(hydroxymethyl)-3,4-bis(oxidanyl)-1-pentyl-pyrrolidin-2-yl]-N-methyl-ethanamide, CALCIUM ION, ... | | Authors: | Bergeron-Brlek, M, Goodwin-Tindall, J, Cekic, N, Varghese, V, Zandberg, W.F, Shan, X, Roth, C, Chan, S, Davies, G.J, Vocadlo, D.J, Britton, R. | | Deposit date: | 2015-08-05 | | Release date: | 2015-11-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Convenient Approach to Stereoisomeric Iminocyclitols: Generation of Potent Brain-Permeable Oga Inhibitors.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5ABE
 
 | | Structure of GH84 with ligand | | Descriptor: | 2-[(2S,3S,4R,5R)-5-(hydroxymethyl)-3,4-bis(oxidanyl)-1-pentyl-pyrrolidin-2-yl]-N-methyl-ethanamide, CALCIUM ION, O-GLCNACASE BT_4395 | | Authors: | Bergeron-Brlek, M, Goodwin-Tindall, J, Cekic, N, Varghese, V, Zandberg, W.F, Shan, X, Roth, C, Chan, S, Davies, G.J, Vocadlo, D.J, Britton, R. | | Deposit date: | 2015-08-05 | | Release date: | 2015-11-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Convenient Approach to Stereoisomeric Iminocyclitols: Generation of Potent Brain-Permeable Oga Inhibitors.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4AEL
 
 | | PDE10A in complex with the inhibitor AZ5 | | Descriptor: | 2-(2'-ETHOXYBIPHENYL-4-YL)-4-HYDROXY-1,6-NAPHTHYRIDINE-3-CARBONITRILE, CAMP AND CAMP-INHIBITED CGMP 3', 5'-CYCLIC PHOSPHODIESTERASE 10A, ... | | Authors: | Bauer, U, Giordanetto, F, Bauer, M, OMahony, G, Johansson, K.E, Knecht, W, Hartleib-Geschwindner, J, Toppner Carlsson, E, Enroth, C, Sjogren, T. | | Deposit date: | 2012-01-11 | | Release date: | 2012-01-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of 4-Hydroxy-1,6-Naphthyridine-3-Carbonitrile Derivatives as Novel Pde10A Inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
8BAL
 
 | |
8BBL
 
 | | SGL a GH20 family sulfoglycosidase | | Descriptor: | Beta-N-acetylhexosaminidase | | Authors: | Dong, M.D, Roth, C.R, Jin, Y.J. | | Deposit date: | 2022-10-13 | | Release date: | 2023-01-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.711 Å) | | Cite: | Mechanistic and Structural Insights into the Specificity and Biological Functions of Bacterial Sulfoglycosidases
Acs Catalysis, 2022
|
|
7DUP
 
 | | Apo structure of wild type Bt4394, a GH20 family sulfoglycosidase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-N-acetylhexosaminidase, CHLORIDE ION, ... | | Authors: | Zhang, Z, He, Y, Jin, Y. | | Deposit date: | 2021-01-10 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Mechanistic and Structural Insights into the Specificity and Biological Functions of Bacterial Sulfoglycosidases
Acs Catalysis, 13, 2023
|
|
7DVA
 
 | | Structure of wild type Bt4394, a GH20 family sulfoglycosidase, in complex with 6S-GlcNAc | | Descriptor: | 2-acetamido-2-deoxy-6-O-sulfo-beta-D-glucopyranose, Beta-N-acetylhexosaminidase, GLYCEROL | | Authors: | Zhang, Z, He, Y, Jin, Y. | | Deposit date: | 2021-01-13 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Mechanistic and Structural Insights into the Specificity and Biological Functions of Bacterial Sulfoglycosidases
Acs Catalysis, 13, 2023
|
|
7DVB
 
 | | D335N variant of Bt4394 in complex with 6SO3-NAG-oxazoline intermediate | | Descriptor: | 2-acetamido-2-deoxy-6-O-sulfo-beta-D-glucopyranose, Beta-N-acetylhexosaminidase, [(3~{a}~{R},5~{R},6~{S},7~{R},7~{a}~{R})-2-methyl-6,7-bis(oxidanyl)-5,6,7,7~{a}-tetrahydro-3~{a}~{H}-pyrano[3,2-d][1,3]oxazol-1-ium-5-yl]methyl sulfate | | Authors: | Zhang, Z, He, Y, Jin, Y. | | Deposit date: | 2021-01-13 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Mechanistic and Structural Insights into the Specificity and Biological Functions of Bacterial Sulfoglycosidases
Acs Catalysis, 13, 2023
|
|
6YJS
 
 | | Crystal structure of MGAT5 (alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase V) luminal domain with a Lys329-Ile345 loop truncation, in complex with biantennary pentasaccharide M592 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose, ... | | Authors: | Wu, L, Darby, J.F, Gilio, A.K, Davies, G.J. | | Deposit date: | 2020-04-04 | | Release date: | 2020-08-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Substrate Engagement and Catalytic Mechanisms of N-Acetylglucosaminyltransferase V
Acs Catalysis, 2020
|
|
6YJT
 
 | | Crystal structure of MGAT5 (alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase V) luminal domain with a Lys329-Ile345 loop truncation, in complex with UDP | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase A, ... | | Authors: | Wu, L, Darby, J.F, Gilio, A.K, Davies, G.J. | | Deposit date: | 2020-04-04 | | Release date: | 2020-08-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Substrate Engagement and Catalytic Mechanisms of N-Acetylglucosaminyltransferase V
Acs Catalysis, 2020
|
|
6YJQ
 
 | | Crystal structure of unliganded MGAT5 (alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase V) luminal domain with a Lys329-Ile345 loop truncation | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase A,Alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase A, ... | | Authors: | Wu, L, Darby, J.F, Gilio, A.K, Davies, G.J. | | Deposit date: | 2020-04-04 | | Release date: | 2020-08-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Substrate Engagement and Catalytic Mechanisms of N-Acetylglucosaminyltransferase V
Acs Catalysis, 2020
|
|
6YJV
 
 | | Crystal structure of unliganded MGAT5 (alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase V) luminal domain with a Lys329-Ile345 loop truncation, in complex with UDP-2-deoxy-2-fluoroglucose and biantennary pentasaccharide M592 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose, ... | | Authors: | Wu, L, Darby, J.F, Gilio, A.K, Davies, G.J. | | Deposit date: | 2020-04-04 | | Release date: | 2020-08-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Substrate Engagement and Catalytic Mechanisms of N-Acetylglucosaminyltransferase V
Acs Catalysis, 2020
|
|
6YJR
 
 | | Crystal structure of unliganded MGAT5 (alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase V) luminal domain. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase A, ... | | Authors: | Wu, L, Darby, J.F, Gilio, A.K, Davies, G.J. | | Deposit date: | 2020-04-04 | | Release date: | 2020-08-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Substrate Engagement and Catalytic Mechanisms of N-Acetylglucosaminyltransferase V
Acs Catalysis, 2020
|
|
