5HV6
 
 | |
5HV3
 
 | |
5HV2
 
 | |
9MS8
 
 | | PTCH1 in complex with Fab6H3 | | Descriptor: | 6H3 Fab heavy chain, 6H3 Fab light chain, Protein patched homolog 1 | | Authors: | Hu, Q, Qi, X, Li, X. | | Deposit date: | 2025-01-09 | | Release date: | 2025-04-02 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Inhibiting hedgehog signal by a patched-1 antibody.
Cell Discov, 11, 2025
|
|
6UM1
 
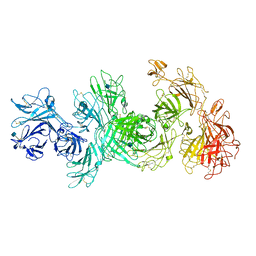 | | Structure of M-6-P/IGFII Receptor at pH 4.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cation-independent mannose-6-phosphate receptor, ... | | Authors: | Wang, R, Qi, X, Li, X. | | Deposit date: | 2019-10-08 | | Release date: | 2020-02-26 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Marked structural rearrangement of mannose 6-phosphate/IGF2 receptor at different pH environments
Sci Adv, 6, 2020
|
|
6UM2
 
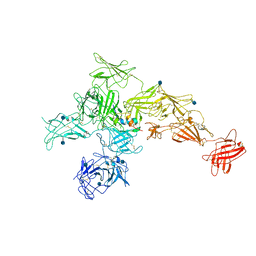 | | Structure of M-6-P/IGFII Receptor and IGFII complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cation-independent mannose-6-phosphate receptor, Insulin-like growth factor II | | Authors: | Wang, R, Qi, X, Li, X. | | Deposit date: | 2019-10-08 | | Release date: | 2020-02-26 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.32 Å) | | Cite: | Marked structural rearrangement of mannose 6-phosphate/IGF2 receptor at different pH environments
Sci Adv, 6, 2020
|
|
8DJM
 
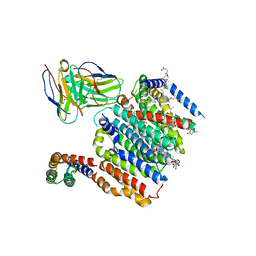 | | HMGCR-UBIAD1 Complex State 1 | | Descriptor: | 3-hydroxy-3-methylglutaryl-coenzyme A reductase, CHOLESTEROL HEMISUCCINATE, Digitonin, ... | | Authors: | Chen, H, Qi, X, Li, X. | | Deposit date: | 2022-07-01 | | Release date: | 2022-08-03 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Regulated degradation of HMG CoA reductase requires conformational changes in sterol-sensing domain.
Nat Commun, 13, 2022
|
|
8DJK
 
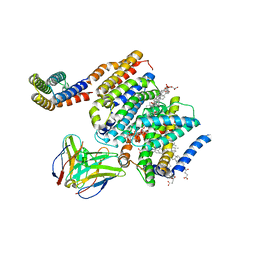 | | HMGCR-UBIAD1 Complex State 2 | | Descriptor: | 3-hydroxy-3-methylglutaryl-coenzyme A reductase, CHOLESTEROL HEMISUCCINATE, Digitonin, ... | | Authors: | Chen, H, Qi, X, Li, X. | | Deposit date: | 2022-06-30 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Regulated degradation of HMG CoA reductase requires conformational changes in sterol-sensing domain.
Nat Commun, 13, 2022
|
|
9BUM
 
 | | Structure of gamma-glutamyl carboxylase (GGCX) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, Vitamin K-dependent gamma-carboxylase | | Authors: | Wang, R, Qi, X. | | Deposit date: | 2024-05-17 | | Release date: | 2025-01-29 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Structure and mechanism of vitamin-K-dependent gamma-glutamyl carboxylase.
Nature, 639, 2025
|
|
9BUX
 
 | | Structure of GGCX-BGP-menaquinone-4 epoxide complex | | Descriptor: | (1aR,7aS)-1a-methyl-7a-[(2E,6E,10E)-3,7,11,15-tetramethylhexadeca-2,6,10,14-tetraen-1-yl]-1a,7a-dihydronaphtho[2,3-b]oxirene-2,7-dione, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, R, Qi, X. | | Deposit date: | 2024-05-17 | | Release date: | 2025-01-29 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structure and mechanism of vitamin-K-dependent gamma-glutamyl carboxylase.
Nature, 639, 2025
|
|
9BUR
 
 | | Structure of GGCX-BGP complex | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, R, Qi, X. | | Deposit date: | 2024-05-17 | | Release date: | 2025-01-29 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structure and mechanism of vitamin-K-dependent gamma-glutamyl carboxylase.
Nature, 639, 2025
|
|
5J6A
 
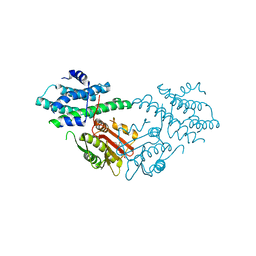 | | Crystal structure of pyruvate dehydrogenase kinase isoform 2 in complex with inhibitor PS46 | | Descriptor: | (3S)-3-amino-4-[4-({2-[(2,4-dihydroxyphenyl)sulfonyl]-2H-isoindol-5-yl}amino)piperidin-1-yl]-4-oxobutanamide, [Pyruvate dehydrogenase (acetyl-transferring)] kinase isozyme 2, mitochondrial | | Authors: | Gui, W.J, Tso, S.C, Chuang, J.L, Wu, C.Y, Qi, X, Wynn, R.M, Chuang, D.T. | | Deposit date: | 2016-04-04 | | Release date: | 2017-01-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.045 Å) | | Cite: | Development of Dihydroxyphenyl Sulfonylisoindoline Derivatives as Liver-Targeting Pyruvate Dehydrogenase Kinase Inhibitors.
J. Med. Chem., 60, 2017
|
|
4MPN
 
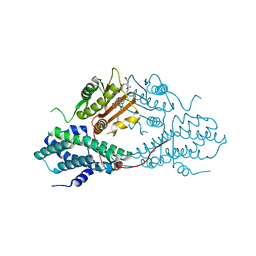 | | Crystal structure of pyruvate dehydrogenase kinase isoform 2 in complex with inhibitor PS10 | | Descriptor: | 2-[(2,4-dihydroxyphenyl)sulfonyl]-2,3-dihydro-1H-isoindole-4,6-diol, L(+)-TARTARIC ACID, [Pyruvate dehydrogenase [lipoamide]] kinase isozyme 2, ... | | Authors: | Gui, W.J, Tso, S.C, Chuang, J.L, Wu, C.Y, Qi, X, Tambar, U.K, Wynn, R.M, Chuang, D.T. | | Deposit date: | 2013-09-13 | | Release date: | 2014-01-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Structure-guided Development of Specific Pyruvate Dehydrogenase Kinase Inhibitors Targeting the ATP-binding Pocket.
J.Biol.Chem., 289, 2014
|
|
4MPE
 
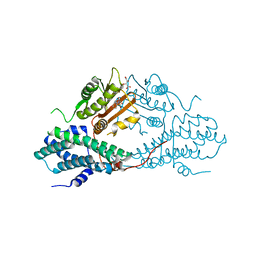 | | Crystal structure of pyruvate dehydrogenase kinase isoform 2 in complex with inhibitor PS8 | | Descriptor: | 4-[(5-hydroxy-1,3-dihydro-2H-isoindol-2-yl)sulfonyl]benzene-1,3-diol, L(+)-TARTARIC ACID, [Pyruvate dehydrogenase [lipoamide]] kinase isozyme 2, ... | | Authors: | Gui, W.J, Tso, S.C, Chuang, J.L, Wu, C.Y, Qi, X, Tambar, U.K, Wynn, R.M, Chuang, D.T. | | Deposit date: | 2013-09-12 | | Release date: | 2014-01-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.953 Å) | | Cite: | Structure-guided Development of Specific Pyruvate Dehydrogenase Kinase Inhibitors Targeting the ATP-binding Pocket.
J.Biol.Chem., 289, 2014
|
|
4MP7
 
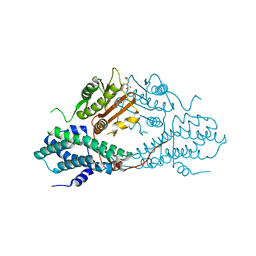 | | Crystal structure of pyruvate dehydrogenase kinase isoform 2 in complex with inhibitor PA7 | | Descriptor: | 4-(1,3-dihydro-2H-isoindol-2-ylcarbonyl)benzene-1,3-diol, L(+)-TARTARIC ACID, [Pyruvate dehydrogenase [lipoamide]] kinase isozyme 2, ... | | Authors: | Gui, W.J, Tso, S.C, Chuang, J.L, Wu, C.Y, Qi, X, Tambar, U.K, Wynn, R.M, Chuang, D.T. | | Deposit date: | 2013-09-12 | | Release date: | 2014-01-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-guided Development of Specific Pyruvate Dehydrogenase Kinase Inhibitors Targeting the ATP-binding Pocket.
J.Biol.Chem., 289, 2014
|
|
4MPC
 
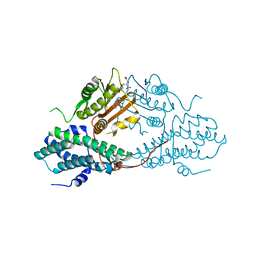 | | Crystal structure of pyruvate dehydrogenase kinase isoform 2 in complex with inhibitor PS2 | | Descriptor: | 4-(isoindolin-2-ylsulfonyl)benzene-1,3-diol, L(+)-TARTARIC ACID, [Pyruvate dehydrogenase [lipoamide]] kinase isozyme 2, ... | | Authors: | Gui, W.J, Tso, S.C, Chuang, J.L, Wu, C.Y, Qi, X, Tambar, U.K, Wynn, R.M, Chuang, D.T. | | Deposit date: | 2013-09-12 | | Release date: | 2014-01-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Structure-guided Development of Specific Pyruvate Dehydrogenase Kinase Inhibitors Targeting the ATP-binding Pocket.
J.Biol.Chem., 289, 2014
|
|
4MP2
 
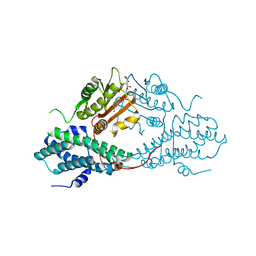 | | Crystal structure of pyruvate dehydrogenase kinase isoform 2 in complex with inhibitor PA1 | | Descriptor: | (5-bromo-2,4-dihydroxyphenyl)(1,3-dihydro-2H-isoindol-2-yl)methanone, L(+)-TARTARIC ACID, [Pyruvate dehydrogenase [lipoamide]] kinase isozyme 2, ... | | Authors: | Gui, W.J, Tso, S.C, Chuang, J.L, Wu, C.Y, Qi, X, Tambar, U.K, Wynn, R.M, Chuang, D.T. | | Deposit date: | 2013-09-12 | | Release date: | 2014-01-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Structure-guided Development of Specific Pyruvate Dehydrogenase Kinase Inhibitors Targeting the ATP-binding Pocket.
J.Biol.Chem., 289, 2014
|
|
4KMA
 
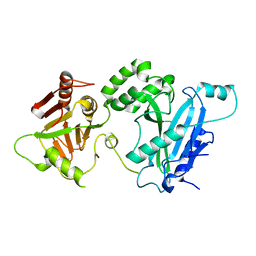 | |
4KMH
 
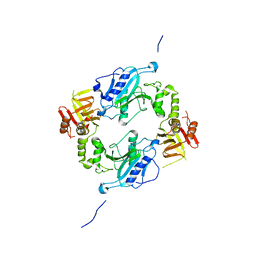 | |
4KM9
 
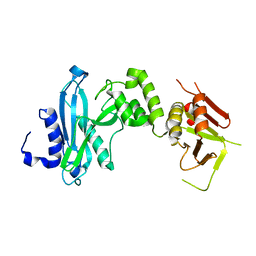 | |
4KMD
 
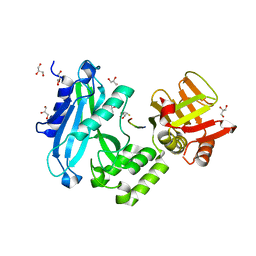 | | Crystal structure of Sufud60-Gli1p | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, GLYCEROL, Sufu, ... | | Authors: | Zhang, Y, Qi, X, Zhang, Z, Wu, G. | | Deposit date: | 2013-05-08 | | Release date: | 2013-11-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insight into the mutual recognition and regulation between Suppressor of Fused and Gli/Ci.
Nat Commun, 4, 2013
|
|
5J71
 
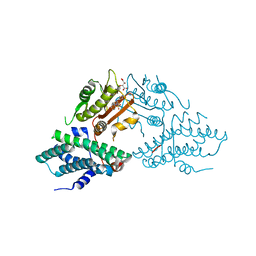 | | Crystal structure of pyruvate dehydrogenase kinase isoform 2 in complex with inhibitor PS35 | | Descriptor: | 4-({5-[(piperidin-4-yl)amino]-1,3-dihydro-2H-isoindol-2-yl}sulfonyl)benzene-1,3-diol, L(+)-TARTARIC ACID, [Pyruvate dehydrogenase (acetyl-transferring)] kinase isozyme 2, ... | | Authors: | Gui, W.J, Tso, S.C, Chuang, J.L, Wu, C.Y, Qi, X, Wynn, R.M, Chuang, D.T. | | Deposit date: | 2016-04-05 | | Release date: | 2017-01-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Development of Dihydroxyphenyl Sulfonylisoindoline Derivatives as Liver-Targeting Pyruvate Dehydrogenase Kinase Inhibitors.
J. Med. Chem., 60, 2017
|
|
4KM8
 
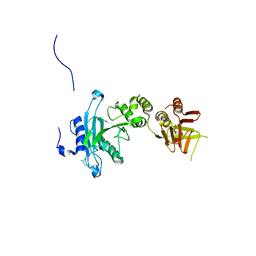 | | Crystal structure of Sufud60 | | Descriptor: | Suppressor of fused homolog | | Authors: | Zhang, Y, Qi, X, Zhang, Z, Wu, G. | | Deposit date: | 2013-05-08 | | Release date: | 2013-11-20 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structural insight into the mutual recognition and regulation between Suppressor of Fused and Gli/Ci.
Nat Commun, 4, 2013
|
|
5X41
 
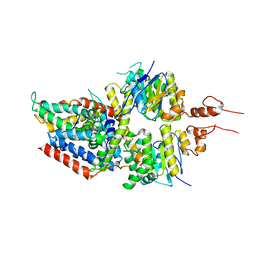 | | 3.5A resolution structure of a cobalt energy-coupling factor transporter using LCP method-CbiMQO | | Descriptor: | Cobalt ABC transporter ATP-binding protein, Cobalt transport protein CbiM, Uncharacterized protein CbiQ | | Authors: | Bao, Z, Qi, X, Zhao, W, Li, D, Zhang, P. | | Deposit date: | 2017-02-09 | | Release date: | 2017-04-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.47 Å) | | Cite: | Structure and mechanism of a group-I cobalt energy coupling factor transporter
Cell Res., 27, 2017
|
|
5X40
 
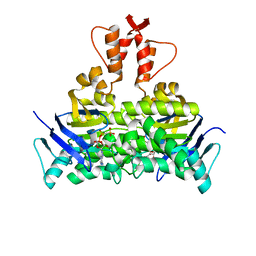 | | Structure of a CbiO dimer bound with AMPPCP | | Descriptor: | Cobalt ABC transporter ATP-binding protein, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Bao, Z, Qi, X, Wang, J, Zhang, P. | | Deposit date: | 2017-02-09 | | Release date: | 2017-04-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure and mechanism of a group-I cobalt energy coupling factor transporter
Cell Res., 27, 2017
|
|
