7D1I
 
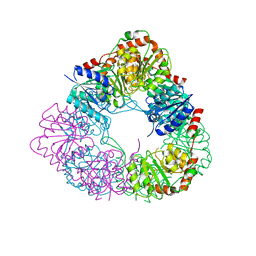 | | Crystal structure of acinetobacter baumannii MurG | | Descriptor: | UDP-N-acetylglucosamine--N-acetylmuramyl-(pentapeptide) pyrophosphoryl-undecaprenol N-acetylglucosamine transferase | | Authors: | Park, H.H, Jeong, k.H. | | Deposit date: | 2020-09-14 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.487 Å) | | Cite: | Putative hexameric glycosyltransferase functional unit revealed by the crystal structure of Acinetobacter baumannii MurG
Iucrj, 8, 2021
|
|
7D27
 
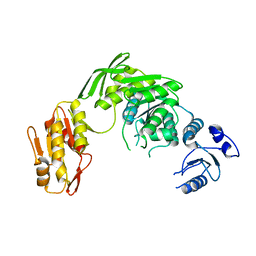 | |
4OM7
 
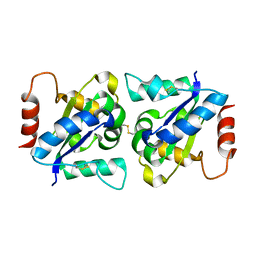 | | Crystal structure of TIR domain of TLR6 | | Descriptor: | Toll-like receptor 6 | | Authors: | Park, H.H, Jang, T.H. | | Deposit date: | 2014-01-26 | | Release date: | 2014-08-06 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Crystal Structure of TIR Domain of TLR6 Reveals Novel Dimeric Interface of TIR-TIR Interaction for Toll-Like Receptor Signaling Pathway.
J.Mol.Biol., 426, 2014
|
|
5YC1
 
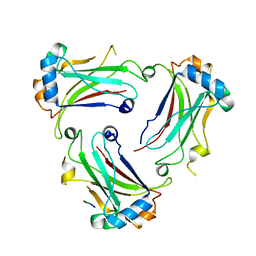 | | TRAF4_GPIb complex | | Descriptor: | GPIb peptide, TNF receptor-associated factor 4 | | Authors: | Park, H.H, Kim, C.M. | | Deposit date: | 2017-09-06 | | Release date: | 2017-10-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.506 Å) | | Cite: | Molecular basis for unique specificity of human TRAF4 for platelets GPIb beta and GPVI
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5ZYN
 
 | | Fumarate reductase | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, Fumarate reductase 2, ... | | Authors: | Park, H.H, Kim, C.M. | | Deposit date: | 2018-05-25 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Molecular basis of maintaining an oxidizing environment under anaerobiosis by soluble fumarate reductase.
Nat Commun, 9, 2018
|
|
5ZTX
 
 | | co-factor free Transaminase | | Descriptor: | 1,2-ETHANEDIOL, transaminase | | Authors: | Park, H.H, Shin, Y.C. | | Deposit date: | 2018-05-05 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural dynamics of the transaminase active site revealed by the crystal structure of a co-factor free omega-transaminase from Vibrio fluvialis JS17
Sci Rep, 8, 2018
|
|
6A8P
 
 | | Transglutaminase 2 mutant G224V in complex with GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Protein-glutamine gamma-glutamyltransferase 2 | | Authors: | Park, H.H, Ha, H.J, Kwon, S. | | Deposit date: | 2018-07-09 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.537 Å) | | Cite: | Structure of natural variant transglutaminase 2 reveals molecular basis of gaining stability and higher activity.
PLoS ONE, 13, 2018
|
|
6IO1
 
 | |
6J52
 
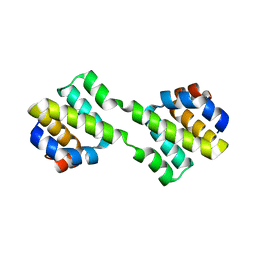 | | Crystal structure of CARD-only protein in frog virus 3 | | Descriptor: | Caspase recruitment domain-only protein | | Authors: | Park, H.H, Kwon, S. | | Deposit date: | 2019-01-10 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.504 Å) | | Cite: | Structural transformation-mediated dimerization of caspase recruitment domain revealed by the crystal structure of CARD-only protein in frog virus 3.
J. Struct. Biol., 205, 2019
|
|
6K8H
 
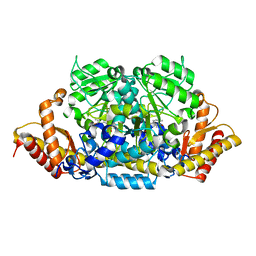 | | Crystal structure of an omega-transaminase from Sphaerobacter thermophilus | | Descriptor: | (5-HYDROXY-4,6-DIMETHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, Aminotransferase class-III | | Authors: | Park, H.H, Kwon, S. | | Deposit date: | 2019-06-12 | | Release date: | 2019-10-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into the enzyme specificity of a novel omega-transaminase from the thermophilic bacterium Sphaerobacter thermophilus.
J.Struct.Biol., 208, 2019
|
|
6KU5
 
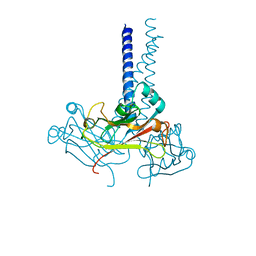 | | Notothenia coriiceps TRAF5 | | Descriptor: | TRAF5 | | Authors: | Park, H.H, Kim, C.M. | | Deposit date: | 2019-08-30 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.299 Å) | | Cite: | Structural and biochemical characterization of TRAF5 from Notothenia coriiceps and its implications in fish immune cell signaling.
Fish Shellfish Immunol., 102, 2020
|
|
6KU6
 
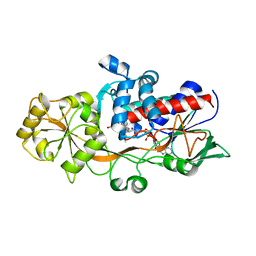 | | OSM1 mutant - R326A | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fumarate reductase 2, SUCCINIC ACID | | Authors: | Park, H.H, Kim, C.M. | | Deposit date: | 2019-08-30 | | Release date: | 2020-07-08 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.007 Å) | | Cite: | Crystal Structure of the Active Site Mutant Form of Soluble Fumarate Reductase, Osm1
Crystals, 2019
|
|
6KZB
 
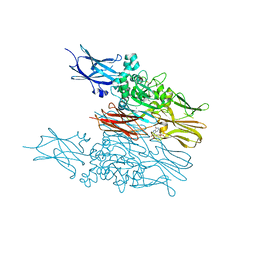 | | Transglutaminase2 complexed with calcium | | Descriptor: | CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, Protein-glutamine gamma-glutamyltransferase 2 | | Authors: | Park, H.H, Kim, C.M. | | Deposit date: | 2019-09-23 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Competitive Binding of Magnesium to Calcium Binding Sites Reciprocally Regulates Transamidase and GTP Hydrolysis Activity of Transglutaminase 2.
Int J Mol Sci, 21, 2020
|
|
6IZ9
 
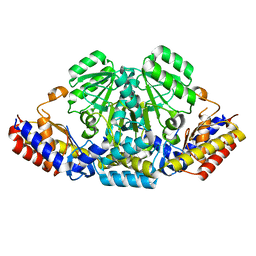 | |
4UZ0
 
 | | Crystal Structure of apoptosis repressor with CARD (ARC) | | Descriptor: | GLYCEROL, NUCLEOLAR PROTEIN 3 | | Authors: | Kim, S.H, Jeong, J.H, Jang, T.H, Kim, Y.G, Park, H.H. | | Deposit date: | 2014-09-04 | | Release date: | 2015-07-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Crystal Structure of Caspase Recruiting Domain (Card) of Apoptosis Repressor with Card (Arc) and its Implication in Inhibition of Apoptosis.
Sci.Rep., 5, 2015
|
|
7XI1
 
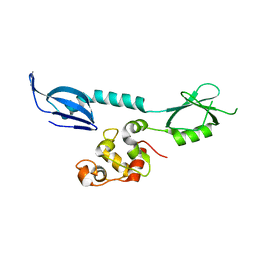 | | AcrIF 24 | | Descriptor: | anti-CRISPR protein AcrIF24 | | Authors: | Kim, G.E, Park, H.H. | | Deposit date: | 2022-04-11 | | Release date: | 2023-03-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Molecular basis of dual anti-CRISPR and auto-regulatory functions of AcrIF24.
Nucleic Acids Res., 50, 2022
|
|
7YHR
 
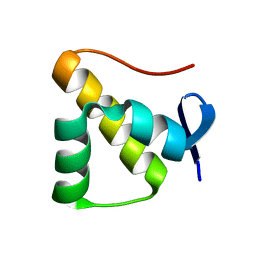 | | Anti-CRISPR protein AcrIC5 | | Descriptor: | Anti-CRISPR protein Type I-C5 | | Authors: | Kang, Y.J, Park, H.H. | | Deposit date: | 2022-07-14 | | Release date: | 2023-05-24 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | High-resolution crystal structure of the anti-CRISPR protein AcrIC5.
Biochem.Biophys.Res.Commun., 625, 2022
|
|
9KJB
 
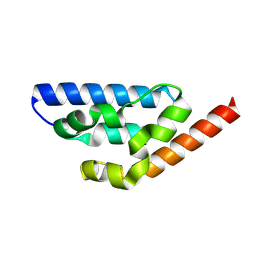 | | Crystal structure of Anti-CRISPR AcrIE7 | | Descriptor: | AcrIE7 | | Authors: | Kim, D.Y, Park, H.H. | | Deposit date: | 2024-11-12 | | Release date: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | AcrIE7 inhibits the CRISPR-Cas system by directly binding to the R-loop single-stranded DNA.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
7YSI
 
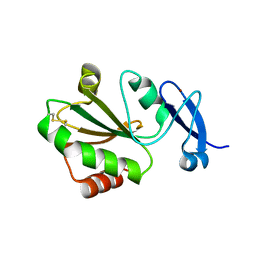 | | Crystal structure of thioredoxin 2 | | Descriptor: | Thiol disulfide reductase thioredoxin, ZINC ION | | Authors: | Chang, Y.J, Park, H.H. | | Deposit date: | 2022-08-12 | | Release date: | 2023-03-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.202 Å) | | Cite: | Comparison of the structure and activity of thioredoxin 2 and thioredoxin 1 from Acinetobacter baumannii.
Iucrj, 10, 2023
|
|
4D2K
 
 | |
5EZ5
 
 | |
1YYF
 
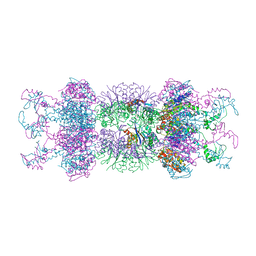 | | Correction of X-ray Intensities from an HslV-HslU co-crystal containing lattice translocation defects | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent hsl protease ATP-binding subunit hslU, ATP-dependent protease hslV | | Authors: | Wang, J, Rho, S.H, Park, H.H, Eom, S.H. | | Deposit date: | 2005-02-24 | | Release date: | 2005-07-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (4.16 Å) | | Cite: | Correction of X-ray intensities from an HslV-HslU co-crystal containing lattice-translocation defects.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
8K2Y
 
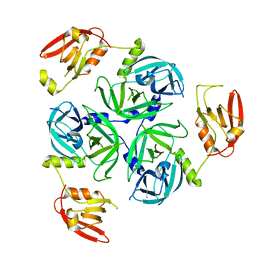 | | Crystal structure of MucD | | Descriptor: | serine endoprotease DegP-like protein MucD | | Authors: | Kim, J.H, Park, H.H. | | Deposit date: | 2023-07-14 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structure of MucD from Pseudomonas syringae revealed N-terminal loop-mediated trimerization of HtrA-like serine protease.
Biochem.Biophys.Res.Commun., 688, 2023
|
|
8JQZ
 
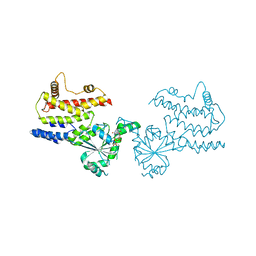 | | Crystal Structure of GppNHp-bound mIRGB10 | | Descriptor: | Immunity-related GTPase family member b10, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Ha, H.J, Park, H.H. | | Deposit date: | 2023-06-15 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural basis of IRGB10 oligomerization by GTP hydrolysis.
Front Immunol, 14, 2023
|
|
8JQY
 
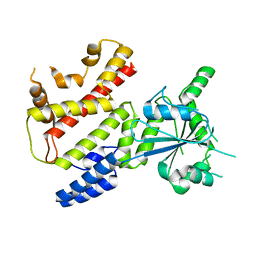 | |
