5D10
 
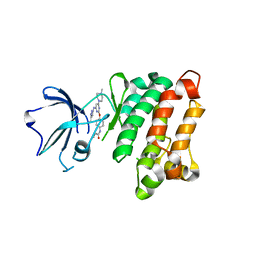 | | Kinase domain of cSrc in complex with RL236 | | Descriptor: | N-[4-({4-(4-methylpiperazin-1-yl)-6-[(5-methyl-1H-pyrazol-3-yl)amino]pyrimidin-2-yl}oxy)phenyl]prop-2-enamide, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Becker, C, Mayer-Wrangowski, S.C, Julian, E, Rauh, D. | | Deposit date: | 2015-08-03 | | Release date: | 2015-09-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Targeting Drug Resistance in EGFR with Covalent Inhibitors: A Structure-Based Design Approach.
J.Med.Chem., 58, 2015
|
|
5D11
 
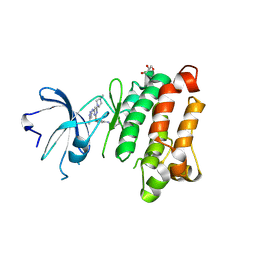 | | Kinase domain of cSrc in complex with RL235 | | Descriptor: | GLYCEROL, N-[3-({4-(4-methylpiperazin-1-yl)-6-[(5-methyl-1H-pyrazol-3-yl)amino]pyrimidin-2-yl}oxy)phenyl]prop-2-enamide, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Becker, C, Gruetter, C, Engel, J, Rauh, D. | | Deposit date: | 2015-08-03 | | Release date: | 2015-09-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Targeting Drug Resistance in EGFR with Covalent Inhibitors: A Structure-Based Design Approach.
J.Med.Chem., 58, 2015
|
|
5D12
 
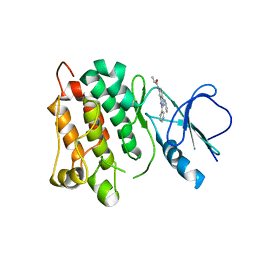 | | Kinase domain of cSrc in complex with RL40 | | Descriptor: | N-[2-phenyl-4-(1H-pyrazol-3-ylamino)quinazolin-7-yl]prop-2-enamide, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Becker, C, Richters, A, Engel, J, Rauh, D. | | Deposit date: | 2015-08-03 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Targeting Drug Resistance in EGFR with Covalent Inhibitors: A Structure-Based Design Approach.
J.Med.Chem., 58, 2015
|
|
6PER
 
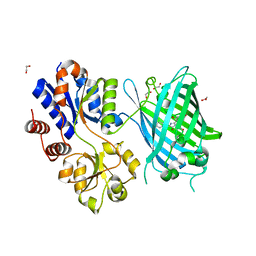 | | Crystal Structure of Ligand-Free iSeroSnFR | | Descriptor: | 1,2-ETHANEDIOL, iSeroSnFR, a soluble, ... | | Authors: | Hartanto, S, Tian, L, Fisher, A.J. | | Deposit date: | 2019-06-20 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Directed Evolution of a Selective and Sensitive Serotonin Sensor via Machine Learning.
Cell, 183, 2020
|
|
7OBH
 
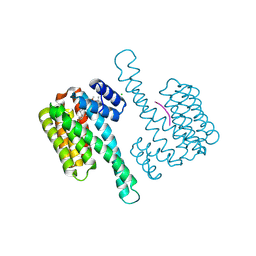 | |
7OBX
 
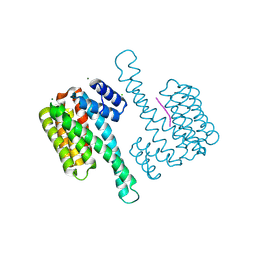 | |
7OBY
 
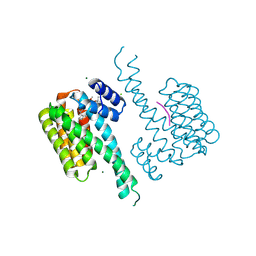 | |
7OBG
 
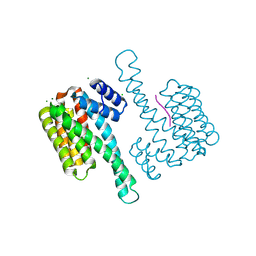 | |
7OBD
 
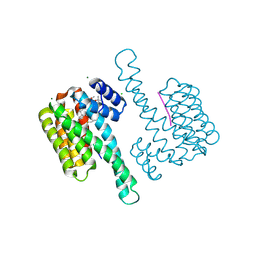 | |
7OBS
 
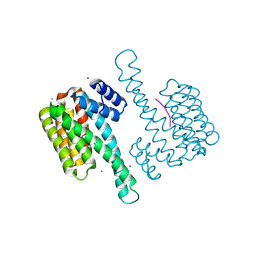 | |
7OBK
 
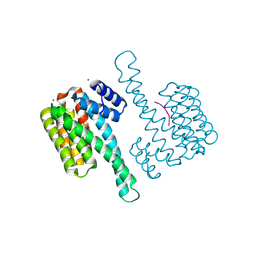 | |
7OB8
 
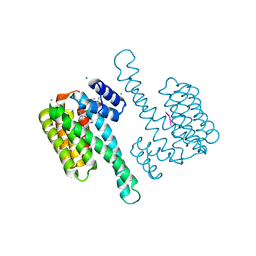 | |
7OBT
 
 | |
7OB5
 
 | |
7OBL
 
 | |
7OBC
 
 | |
6QH5
 
 | | AP2 clathrin adaptor mu2T156-phosphorylated core in closed conformation | | Descriptor: | AP-2 complex subunit alpha, AP-2 complex subunit beta, AP-2 complex subunit mu, ... | | Authors: | Wrobel, A.G, Owen, D.J, McCoy, A.J, Evans, P.R. | | Deposit date: | 2019-01-15 | | Release date: | 2019-09-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Temporal Ordering in Endocytic Clathrin-Coated Vesicle Formation via AP2 Phosphorylation.
Dev.Cell, 50, 2019
|
|
6QH7
 
 | | AP2 clathrin adaptor mu2T156-phosphorylated core with two cargo peptides in open+ conformation | | Descriptor: | ADAPTOR-RELATED PROTEIN COMPLEX 2, MU 2 SUBUNIT, C-TERMINAL DOMAIN, ... | | Authors: | Wrobel, A.G, Owen, D.J, McCoy, A.J, Evans, P.R. | | Deposit date: | 2019-01-15 | | Release date: | 2019-09-04 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Temporal Ordering in Endocytic Clathrin-Coated Vesicle Formation via AP2 Phosphorylation.
Dev.Cell, 50, 2019
|
|
6QH6
 
 | | AP2 clathrin adaptor core with two cargo peptides in open+ conformation | | Descriptor: | AP-2 complex subunit alpha, AP-2 complex subunit beta, AP-2 complex subunit mu, ... | | Authors: | Wrobel, A.G, Owen, D.J, McCoy, A.J, Evans, P.R. | | Deposit date: | 2019-01-15 | | Release date: | 2019-09-04 | | Last modified: | 2022-12-07 | | Method: | X-RAY DIFFRACTION (5 Å) | | Cite: | Temporal Ordering in Endocytic Clathrin-Coated Vesicle Formation via AP2 Phosphorylation.
Dev.Cell, 50, 2019
|
|
6RH5
 
 | | Solution structure and 1H, 13C and 15N chemical shift assignments for NECAP1 PHear domain | | Descriptor: | Adaptin ear-binding coat-associated protein 1 | | Authors: | Owen, D.J, Neuhaus, D, Yang, J.-C, Herrmann, T. | | Deposit date: | 2019-04-18 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Temporal Ordering in Endocytic Clathrin-Coated Vesicle Formation via AP2 Phosphorylation.
Dev.Cell, 50, 2019
|
|
6RH6
 
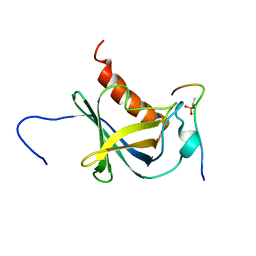 | | Solution structure and 1H, 13C and 15N chemical shift assignments for the complex of NECAP1 PHear domain with phosphorylated AP2 mu2 148-163 | | Descriptor: | AP-2 complex subunit mu, Adaptin ear-binding coat-associated protein 1 | | Authors: | Owen, D.J, Neuhaus, D, Yang, J.-C, Herrmann, T. | | Deposit date: | 2019-04-18 | | Release date: | 2019-09-04 | | Method: | SOLUTION NMR | | Cite: | Temporal Ordering in Endocytic Clathrin-Coated Vesicle Formation via AP2 Phosphorylation.
Dev.Cell, 50, 2019
|
|
3PZ1
 
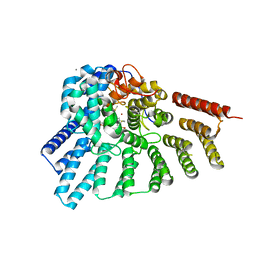 | | Crystal structure of RabGGTase(DELTA LRR; DELTA IG) in Complex with BMS3 | | Descriptor: | (3R)-3-benzyl-4-[(4-methoxyphenyl)sulfonyl]-1-[(1-methyl-1H-imidazol-5-yl)methyl]-2,3,4,5-tetrahydro-1H-1,4-benzodiazepine-7-carbonitrile, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Guo, Z, Bon, R.S, Stigter, E.A, Waldmann, H, Alexandrov, K, Blankenfeldt, W, Goody, R.S. | | Deposit date: | 2010-12-14 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-Guided Development of Selective RabGGTase Inhibitors.
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
3PZ3
 
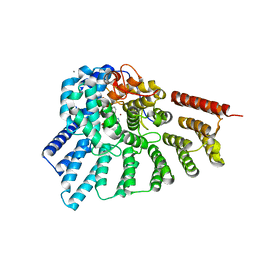 | | Crystal structure of RabGGTase(DELTA LRR; DELTA IG) in Complex with BMS-analogue 14 | | Descriptor: | 4-({(3R)-7-(5-formylfuran-2-yl)-4-[(4-methoxyphenyl)sulfonyl]-1-[(1-methyl-1H-imidazol-5-yl)methyl]-2,3,4,5-tetrahydro-1H-1,4-benzodiazepin-3-yl}methyl)phenyl benzylcarbamate, CALCIUM ION, Geranylgeranyl transferase type-2 subunit alpha, ... | | Authors: | Guo, Z, Bon, R.S, Stigter, E.A, Waldmann, H, Alexandrov, K, Blankenfeldt, W, Goody, R.S. | | Deposit date: | 2010-12-14 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Guided Development of Selective RabGGTase Inhibitors.
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
3PZ2
 
 | | Crystal structure of RabGGTase(DELTA LRR; DELTA IG) in Complex with BMS3 and lipid substrate GGPP | | Descriptor: | (3R)-3-benzyl-4-[(4-methoxyphenyl)sulfonyl]-1-[(1-methyl-1H-imidazol-5-yl)methyl]-2,3,4,5-tetrahydro-1H-1,4-benzodiazepine-7-carbonitrile, CALCIUM ION, GERANYLGERANYL DIPHOSPHATE, ... | | Authors: | Guo, Z, Bon, R.S, Stigter, E.A, Waldmann, H, Alexandrov, K, Blankenfeldt, W, Goody, R.S. | | Deposit date: | 2010-12-14 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure-Guided Development of Selective RabGGTase Inhibitors.
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
3PZ4
 
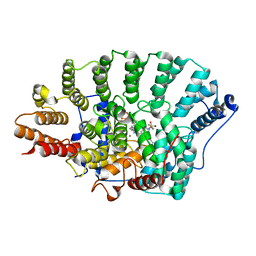 | | Crystal structure of FTase(ALPHA-subunit; BETA-subunit DELTA C10) in complex with BMS3 and lipid substrate FPP | | Descriptor: | (3R)-3-benzyl-4-[(4-methoxyphenyl)sulfonyl]-1-[(1-methyl-1H-imidazol-5-yl)methyl]-2,3,4,5-tetrahydro-1H-1,4-benzodiazepine-7-carbonitrile, FARNESYL DIPHOSPHATE, Protein farnesyltransferase subunit beta, ... | | Authors: | Guo, Z, Bon, R.S, Stigter, E.A, Waldmann, H, Alexandrov, K, Blankenfeldt, W, Goody, R.S. | | Deposit date: | 2010-12-14 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Guided Development of Selective RabGGTase Inhibitors.
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
