1VZE
 
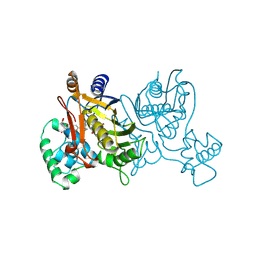 | | L. CASEI THYMIDYLATE SYNTHASE MUTANT E60Q TERNARY COMPLEX WITH DUMP AND CB3717 | | Descriptor: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, THYMIDYLATE SYNTHASE | | Authors: | Birdsall, D.L, Finer-Moore, J, Stroud, R.M. | | Deposit date: | 1996-09-18 | | Release date: | 1997-03-12 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The separate effects of E60Q in Lactobacillus casei thymidylate synthase delineate between mechanisms for formation of intermediates in catalysis.
Protein Eng., 11, 1998
|
|
1VZD
 
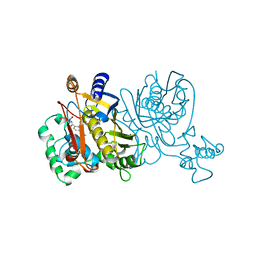 | | L. CASEI THYMIDYLATE SYNTHASE MUTANT E60Q TERNARY COMPLEX WITH FDUMP AND CB3717 | | Descriptor: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, 5-FLUORO-2'-DEOXYURIDINE-5'-MONOPHOSPHATE, THYMIDYLATE SYNTHASE | | Authors: | Birdsall, D.L, Finer-Moore, J, Stroud, R.M. | | Deposit date: | 1996-09-18 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The separate effects of E60Q in Lactobacillus casei thymidylate synthase delineate between mechanisms for formation of intermediates in catalysis.
Protein Eng., 11, 1998
|
|
1VZA
 
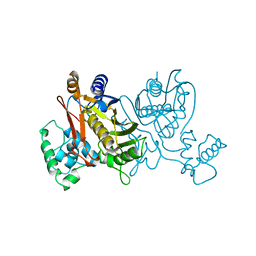 | |
1VZC
 
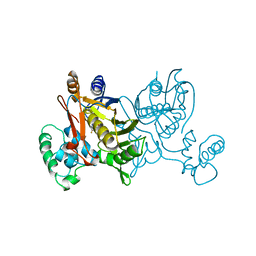 | |
1NCE
 
 | | Crystal structure of a ternary complex of E. coli thymidylate synthase D169C with dUMP and the antifolate CB3717 | | Descriptor: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Thymidylate synthase | | Authors: | Birdsall, D.L, Finer-Moore, J, Stroud, R.M. | | Deposit date: | 2002-12-05 | | Release date: | 2002-12-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The only active mutant of thymidylate synthase D169, a residue far from the
site of methyl transfer, demonstrates the exquisite nature of enzyme
specificity.
Protein Eng., 16, 2003
|
|
1NJB
 
 | | THYMIDYLATE SYNTHASE | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, THYMIDYLATE SYNTHASE | | Authors: | Finer-Moore, J, Stroud, R.M. | | Deposit date: | 1996-01-23 | | Release date: | 1996-10-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Partitioning roles of side chains in affinity, orientation, and catalysis with structures for mutant complexes: asparagine-229 in thymidylate synthase.
Biochemistry, 35, 1996
|
|
1NJA
 
 | | THYMIDYLATE SYNTHASE, MUTATION, N229C WITH 2'-DEOXYCYTIDINE 5'-MONOPHOSPHATE (DCMP) | | Descriptor: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, THYMIDYLATE SYNTHASE | | Authors: | Finer-Moore, J, Stroud, R.M. | | Deposit date: | 1996-01-23 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Partitioning roles of side chains in affinity, orientation, and catalysis with structures for mutant complexes: asparagine-229 in thymidylate synthase.
Biochemistry, 35, 1996
|
|
1NJC
 
 | | THYMIDYLATE SYNTHASE, MUTATION, N229D WITH 2'-DEOXYCYTIDINE 5'-MONOPHOSPHATE (DCMP) | | Descriptor: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, THYMIDYLATE SYNTHASE | | Authors: | Finer-Moore, J, Stroud, R.M. | | Deposit date: | 1996-01-23 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Partitioning roles of side chains in affinity, orientation, and catalysis with structures for mutant complexes: asparagine-229 in thymidylate synthase.
Biochemistry, 35, 1996
|
|
1NJD
 
 | | THYMIDYLATE SYNTHASE, MUTATION, N229D WITH 2'-DEOXYURIDINE 5'-MONOPHOSPHATE (DUMP) | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, THYMIDYLATE SYNTHASE | | Authors: | Finer-Moore, J, Stroud, R.M. | | Deposit date: | 1996-01-23 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Partitioning roles of side chains in affinity, orientation, and catalysis with structures for mutant complexes: asparagine-229 in thymidylate synthase.
Biochemistry, 35, 1996
|
|
2TRM
 
 | |
1BTX
 
 | |
1BTW
 
 | |
1BTZ
 
 | |
1BTY
 
 | |
1NJE
 
 | |
4FGT
 
 | | Allosteric peptidic inhibitor of human thymidylate synthase that stabilizes inactive conformation of the enzyme. | | Descriptor: | CG peptide, SULFATE ION, Thymidylate synthase | | Authors: | Tochowicz, A, Finer-Moore, J, Stroud, R.M, Costi, M.P. | | Deposit date: | 2012-06-04 | | Release date: | 2013-03-06 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Alanine mutants of the interface residues of human thymidylate synthase decode key features of the binding mode of allosteric anticancer peptides.
J.Med.Chem., 58, 2015
|
|
1P8D
 
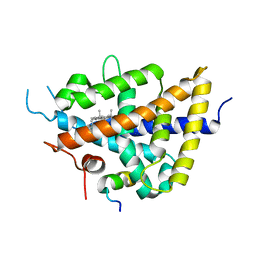 | | X-Ray Crystal Structure of LXR Ligand Binding Domain with 24(S),25-epoxycholesterol | | Descriptor: | 17-[3-(3,3-DIMETHYL-OXIRANYL)-1-METHYL-PROPYL]-10,13-DIMETHYL-2,3,4,7,8,9,10,11,12,13,14,15,16,17-TETRADECAHYDRO-1H-CYC LOPENTA[A]PHENANTHREN-3-OL, Oxysterols receptor LXR-beta, nuclear receptor coactivator 1 isoform 3 | | Authors: | Williams, S, Bledsoe, R.K, Collins, J.L, Boggs, S, Lambert, M.H, Miller, A.B, Moore, J, McKee, D.D, Moore, L, Nichols, J, Parks, D, Watson, M, Wisely, B, Willson, T.M. | | Deposit date: | 2003-05-06 | | Release date: | 2003-07-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray crystal structure of the liver X receptor beta ligand binding domain: regulation by
a histidine-tryptophan switch.
J.Biol.Chem., 278, 2003
|
|
2GCY
 
 | |
5CD1
 
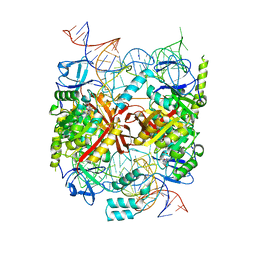 | | Structure of an asymmetric tetramer of human tRNA m1A58 methyltransferase in a complex with SAH and tRNA3Lys | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SODIUM ION, tRNA (adenine(58)-N(1))-methyltransferase catalytic subunit TRMT61A, ... | | Authors: | Finer-Moore, J, Czudnochowski, N, O'Connell III, J.D, Wang, A.L, Stroud, R.M. | | Deposit date: | 2015-07-02 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Crystal Structure of the Human tRNA m(1)A58 Methyltransferase-tRNA3(Lys) Complex: Refolding of Substrate tRNA Allows Access to the Methylation Target.
J.Mol.Biol., 427, 2015
|
|
5CCX
 
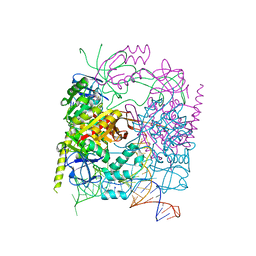 | | Structure of the product complex of tRNA m1A58 methyltransferase with tRNA3Lys as substrate | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SODIUM ION, tRNA (adenine(58)-N(1))-methyltransferase catalytic subunit TRMT61A, ... | | Authors: | Finer-Moore, J, Czudnochowski, N, O'Connell III, J.D, Wang, A.L, Stroud, R.M. | | Deposit date: | 2015-07-02 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Human tRNA m(1)A58 Methyltransferase-tRNA3(Lys) Complex: Refolding of Substrate tRNA Allows Access to the Methylation Target.
J.Mol.Biol., 427, 2015
|
|
5CCB
 
 | | Crystal structure of human m1A58 methyltransferase in a complex with tRNA3Lys and SAH | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SODIUM ION, tRNA (adenine(58)-N(1))-methyltransferase catalytic subunit TRMT61A, ... | | Authors: | Finer-Moore, J, Czudnochowski, N, O'Connell III, J.D, Wang, A.L, Stroud, R.M. | | Deposit date: | 2015-07-01 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Human tRNA m(1)A58 Methyltransferase-tRNA3(Lys) Complex: Refolding of Substrate tRNA Allows Access to the Methylation Target.
J.Mol.Biol., 427, 2015
|
|
2OML
 
 | | crystal structure of E. coli pseudouridine synthase RluE | | Descriptor: | Ribosomal large subunit pseudouridine synthase E, SULFATE ION | | Authors: | Pan, H, Ho, J.D, Stroud, R.M, Finer-Moore, J. | | Deposit date: | 2007-01-22 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The Crystal Structure of E. coli rRNA Pseudouridine Synthase RluE.
J.Mol.Biol., 367, 2007
|
|
2OLW
 
 | | Crystal Structure of E. coli pseudouridine synthase RluE | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ACETIC ACID, Ribosomal large subunit pseudouridine synthase E, ... | | Authors: | Pan, H, Ho, J.D, Stroud, R.M, Finer-Moore, J. | | Deposit date: | 2007-01-19 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Crystal Structure of E. coli rRNA Pseudouridine Synthase RluE.
J.Mol.Biol., 367, 2007
|
|
3FBV
 
 | | Crystal structure of the oligomer formed by the kinase-ribonuclease domain of Ire1 | | Descriptor: | N~2~-1H-benzimidazol-5-yl-N~4~-(3-cyclopropyl-1H-pyrazol-5-yl)pyrimidine-2,4-diamine, Serine/threonine-protein kinase/endoribonuclease IRE1 | | Authors: | Korennykh, A.V, Egea, P.F, Korostelev, A.A, Finer-Moore, J, Zhang, C, Shokat, K.M, Stroud, R.M, Walter, P. | | Deposit date: | 2008-11-19 | | Release date: | 2008-12-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The unfolded protein response signals through high-order assembly of Ire1.
Nature, 457, 2009
|
|
1SJS
 
 | |
