1STZ
 
 | | Crystal structure of a hypothetical protein at 2.2 A resolution | | Descriptor: | Heat-inducible transcription repressor hrcA homolog | | Authors: | Liu, J, Adams, P.D, Shin, D.-H, Huang, C, Yokota, H, Jancarik, J, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-03-25 | | Release date: | 2004-08-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a heat-inducible transcriptional repressor HrcA from Thermotoga maritima: structural insight into DNA binding and dimerization.
J.Mol.Biol., 350, 2005
|
|
6ZVS
 
 | | C12 symmetry: Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily. | | Descriptor: | Vipp1 | | Authors: | Liu, J, Tassinari, M, Souza, D.P, Naskar, S, Noel, J.K, Bohuszewicz, O, Buck, M, Williams, T.A, Baum, B, Low, H.H. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily.
Cell, 184, 2021
|
|
6ZW6
 
 | | C16 symmetry: Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily. | | Descriptor: | vipp1 | | Authors: | Liu, J, Tassinari, M, Souza, D.P, Naskar, S, Noel, J.K, Bohuszewicz, O, Buck, M, Williams, T.A, Baum, B, Low, H.H. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily.
Cell, 184, 2021
|
|
6ZW7
 
 | | C17 symmetry: Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily. | | Descriptor: | vipp1 | | Authors: | Liu, J, Tassinari, M, Souza, D.P, Naskar, S, Noel, J.K, Bohuszewicz, O, Buck, M, Williams, T.A, Baum, B, Low, H.H. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (9.4 Å) | | Cite: | Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily.
Cell, 184, 2021
|
|
1SU0
 
 | | Crystal structure of a hypothetical protein at 2.3 A resolution | | Descriptor: | NifU like protein IscU, ZINC ION | | Authors: | Liu, J, Oganesyan, N, Shin, D.-H, Jancarik, J, Pufan, R, Yokota, H, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-03-25 | | Release date: | 2004-08-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural characterization of an iron-sulfur cluster assembly protein IscU in a zinc-bound form.
Proteins, 59, 2005
|
|
6V51
 
 | | Spin-labeled T4 Lysozyme (9/131FnbY)-(4-Amino-TEMPO) | | Descriptor: | 4-amino-2,2,6,6-tetramethylpiperidin-1-ol, Endolysin | | Authors: | Liu, J, Morizumi, T, Ou, W.L, Wang, L, Ernst, O.P. | | Deposit date: | 2019-12-02 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Genetically Encoded Quinone Methides Enabling Rapid, Site-Specific, and Photocontrolled Protein Modification with Amine Reagents.
J.Am.Chem.Soc., 142, 2020
|
|
8HOG
 
 | | Crystal structure of Bcl-2 in complex with sonrotoclax | | Descriptor: | Apoptosis regulator Bcl-2, ~{N}-[4-[(4-methyl-4-oxidanyl-cyclohexyl)methylamino]-3-nitro-phenyl]sulfonyl-4-[2-[(2~{S})-2-(2-propan-2-ylphenyl)pyrrolidin-1-yl]-7-azaspiro[3.5]nonan-7-yl]-2-(1~{H}-pyrrolo[2,3-b]pyridin-5-yloxy)benzamide | | Authors: | Liu, J, Xu, M, Feng, Y, Hong, Y, Liu, Y. | | Deposit date: | 2022-12-10 | | Release date: | 2024-01-17 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Sonrotoclax overcomes BCL2 G101V mutation-induced venetoclax resistance in preclinical models of hematologic malignancy.
Blood, 143, 2024
|
|
8HOH
 
 | | Crystal structure of Bcl-2 G101V in complex with sonrotoclax | | Descriptor: | Apoptosis regulator Bcl-2, ~{N}-[4-[(4-methyl-4-oxidanyl-cyclohexyl)methylamino]-3-nitro-phenyl]sulfonyl-4-[2-[(2~{S})-2-(2-propan-2-ylphenyl)pyrrolidin-1-yl]-7-azaspiro[3.5]nonan-7-yl]-2-(1~{H}-pyrrolo[2,3-b]pyridin-5-yloxy)benzamide | | Authors: | Liu, J, Xu, M, Feng, Y, Hong, Y, Liu, Y. | | Deposit date: | 2022-12-10 | | Release date: | 2024-01-17 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Sonrotoclax overcomes BCL2 G101V mutation-induced venetoclax resistance in preclinical models of hematologic malignancy.
Blood, 143, 2024
|
|
8HOI
 
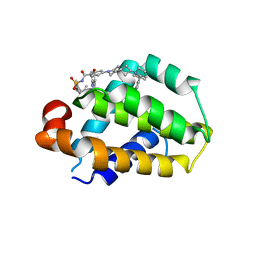 | | Crystal structure of Bcl-2 D103Y in complex with sonrotoclax | | Descriptor: | Apoptosis regulator Bcl-2, FORMIC ACID, ~{N}-[4-[(4-methyl-4-oxidanyl-cyclohexyl)methylamino]-3-nitro-phenyl]sulfonyl-4-[2-[(2~{S})-2-(2-propan-2-ylphenyl)pyrrolidin-1-yl]-7-azaspiro[3.5]nonan-7-yl]-2-(1~{H}-pyrrolo[2,3-b]pyridin-5-yloxy)benzamide | | Authors: | Liu, J, Xu, M, Feng, Y, Hong, Y, Liu, Y. | | Deposit date: | 2022-12-10 | | Release date: | 2024-01-17 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Sonrotoclax overcomes BCL2 G101V mutation-induced venetoclax resistance in preclinical models of hematologic malignancy.
Blood, 143, 2024
|
|
1JPX
 
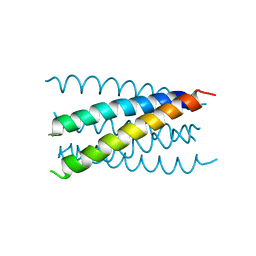 | | Mutation that destabilize the gp41 core: determinants for stabilizing the SIV/CPmac envelope glycoprotein complex. Wild type. | | Descriptor: | gp41 envelope protein | | Authors: | Liu, J, Wang, S, LaBranche, C.C, Hoxie, J.A, Lu, M. | | Deposit date: | 2001-08-03 | | Release date: | 2002-04-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mutations that destabilize the gp41 core are determinants for stabilizing the simian immunodeficiency virus-CPmac envelope glycoprotein complex.
J.Biol.Chem., 277, 2002
|
|
2DFS
 
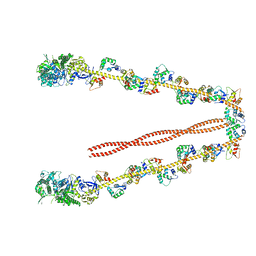 | | 3-D structure of Myosin-V inhibited state | | Descriptor: | Calmodulin, Myosin-5A | | Authors: | Liu, J, Taylor, D.W, Krementsova, E.B, Trybus, K.M, Taylor, K.A. | | Deposit date: | 2006-03-03 | | Release date: | 2006-04-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (24 Å) | | Cite: | Three-dimensional structure of the myosin V inhibited state by cryoelectron tomography
Nature, 442, 2006
|
|
8PE9
 
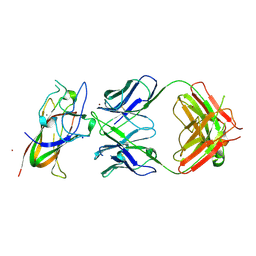 | | Complex between DDR1 DS-like domain and PRTH-101 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Liu, J, Chiang, H, Xiong, W, Laurent, V, Griffiths, S.C, Duelfer, J, Deng, H, Sun, X, Yin, Y.W, Li, W, Audoly, L.P, An, Z, Schuerpf, T, Li, R, Zhang, N. | | Deposit date: | 2023-06-13 | | Release date: | 2023-06-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.152 Å) | | Cite: | A highly selective humanized DDR1 mAb reverses immune exclusion by disrupting collagen fiber alignment in breast cancer.
J Immunother Cancer, 11, 2023
|
|
3I6G
 
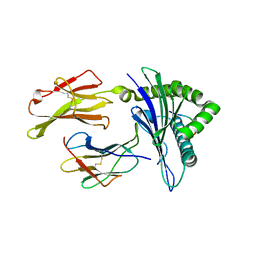 | | Newly identified epitope Mn2 from SARS-CoV M protein complexed withHLA-A*0201 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | Authors: | Liu, J. | | Deposit date: | 2009-07-07 | | Release date: | 2010-06-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | The membrane protein of severe acute respiratory syndrome coronavirus acts as a dominant immunogen revealed by a clustering region of novel functionally and structurally defined cytotoxic T-lymphocyte epitopes.
J.INFECT.DIS., 202, 2010
|
|
6WW4
 
 | | Crystal structure of HERC2 ZZ domain in complex with histone H3 tail | | Descriptor: | GLYCEROL, Histone H3.1,E3 ubiquitin-protein ligase HERC2, ZINC ION | | Authors: | Liu, J, Vann, K.R, Kutateladze, T.G. | | Deposit date: | 2020-05-07 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.252 Å) | | Cite: | Structural Insight into Binding of the ZZ Domain of HERC2 to Histone H3 and SUMO1.
Structure, 28, 2020
|
|
3J3R
 
 | | Structural dynamics of the MecA-ClpC complex revealed by cryo-EM | | Descriptor: | Adapter protein MecA 1, Negative regulator of genetic competence ClpC/MecB | | Authors: | Liu, J, Mei, Z, Li, N, Qi, Y, Xu, Y, Shi, Y, Wang, F, Lei, J, Gao, N. | | Deposit date: | 2013-04-18 | | Release date: | 2013-05-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (9.4 Å) | | Cite: | Structural dynamics of the MecA-ClpC complex: a type II AAA+ protein unfolding machine
J.Biol.Chem., 288, 2013
|
|
3J3T
 
 | | Structural dynamics of the MecA-ClpC complex revealed by cryo-EM | | Descriptor: | Adapter protein MecA 1, Negative regulator of genetic competence ClpC/MecB | | Authors: | Liu, J, Mei, Z, Li, N, Qi, Y, Xu, Y, Shi, Y, Wang, F, Lei, J, Gao, N. | | Deposit date: | 2013-04-18 | | Release date: | 2013-05-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Structural dynamics of the MecA-ClpC complex: a type II AAA+ protein unfolding machine.
J.Biol.Chem., 288, 2013
|
|
1JQ0
 
 | | Mutation that destabilize the gp41 core: determinants for stabilizing the SIV/CPmac envelope glycoprotein complex. Mutant structure. | | Descriptor: | gp41 envelope protein | | Authors: | Liu, J, Wang, S, LaBranche, C.C, Hoxie, J.A, Lu, M. | | Deposit date: | 2001-08-03 | | Release date: | 2002-04-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mutations that destabilize the gp41 core are determinants for stabilizing the simian immunodeficiency virus-CPmac envelope glycoprotein complex.
J.Biol.Chem., 277, 2002
|
|
3J3U
 
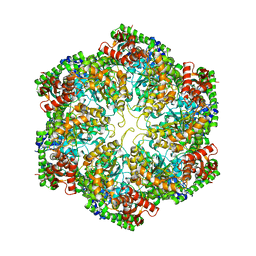 | | Structural dynamics of the MecA-ClpC complex revealed by cryo-EM | | Descriptor: | Adapter protein MecA 1, Negative regulator of genetic competence ClpC/MecB | | Authors: | Liu, J, Mei, Z, Li, N, Qi, Y, Xu, Y, Shi, Y, Wang, F, Lei, J, Gao, N. | | Deposit date: | 2013-04-18 | | Release date: | 2013-05-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Structural dynamics of the MecA-ClpC complex: a type II AAA+ protein unfolding machine.
J.Biol.Chem., 288, 2013
|
|
3J3S
 
 | | Structural dynamics of the MecA-ClpC complex revealed by cryo-EM | | Descriptor: | Adapter protein MecA 1, Negative regulator of genetic competence ClpC/MecB | | Authors: | Liu, J, Mei, Z, Li, N, Qi, Y, Xu, Y, Shi, Y, Wang, F, Lei, J, Gao, N. | | Deposit date: | 2013-04-18 | | Release date: | 2013-05-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Structural dynamics of the MecA-ClpC complex: a type II AAA+ protein unfolding machine.
J.Biol.Chem., 288, 2013
|
|
1SJJ
 
 | |
1FNN
 
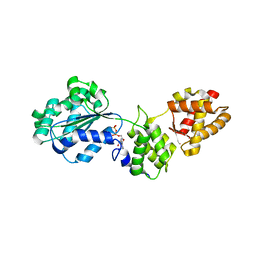 | | CRYSTAL STRUCTURE OF CDC6P FROM PYROBACULUM AEROPHILUM | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CELL DIVISION CONTROL PROTEIN 6, MAGNESIUM ION | | Authors: | Liu, J, Smith, C.L, DeRyckere, D, DeAngelis, K, Martin, G.S, Berger, J.M. | | Deposit date: | 2000-08-22 | | Release date: | 2000-10-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and function of Cdc6/Cdc18: implications for origin recognition and checkpoint control.
Mol.Cell, 6, 2000
|
|
2WR0
 
 | | Structures of influenza H2 Hemagglutinins | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HEMAGGLUTININ, ... | | Authors: | Liu, J, Stevens, D, Haire, L, Coombs, P, Russell, R, Gamblin, S, Skehel, J. | | Deposit date: | 2009-08-28 | | Release date: | 2009-09-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | From the Cover: Structures of Receptor Complexes Formed by Hemagglutinins from the Asian Influenza Pandemic of 1957.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1F23
 
 | | CONTRIBUTION OF A BURIED HYDROGEN BOND TO HIV-1 ENVELOPE GLYCOPROTEIN STRUCTURE AND FUNCTION | | Descriptor: | TRANSMEMBRANE GLYCOPROTEIN | | Authors: | Liu, J, Shu, W, Fagan, M, Nunberg, J.H, Lu, M. | | Deposit date: | 2000-05-23 | | Release date: | 2001-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and functional analysis of the HIV gp41 core containing an Ile573 to Thr substitution: implications for membrane fusion.
Biochemistry, 40, 2001
|
|
5YJ8
 
 | |
5GP0
 
 | | Crystal structure of geraniol-NUDX1 complex | | Descriptor: | GERANYL DIPHOSPHATE, GLYCEROL, Nudix hydrolase 1 | | Authors: | Liu, J, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-07-30 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Structural Insights into the Substrate Recognition Mechanism of Arabidopsis GPP-Bound NUDX1 for Noncanonical Monoterpene Biosynthesis.
Mol Plant, 11, 2018
|
|
