4RWB
 
 | | Racemic influenza M2-TM crystallized from monoolein lipidic cubic phase | | Descriptor: | Matrix protein 2, [(Z)-octadec-9-enyl] (2R)-2,3-bis(oxidanyl)propanoate | | Authors: | Mortenson, D.E, Steinkruger, J.D, Kreitler, D.F, Gellman, S.H, Forest, K.T. | | Deposit date: | 2014-12-02 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution structures of a heterochiral coiled coil.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
8D52
 
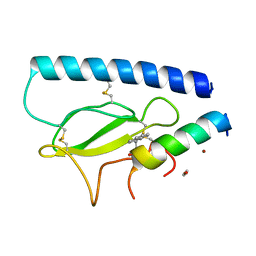 | | Parathyroid hormone 1 receptor extracellular domain complexed with a peptide ligand containing (2-naphthyl)-beta-3-homoalanine | | Descriptor: | 1,2-ETHANEDIOL, PTHrP[1-36], Parathyroid hormone/parathyroid hormone-related peptide receptor, ... | | Authors: | Yu, Z, Kreitler, D.F, Gellman, S.H. | | Deposit date: | 2022-06-03 | | Release date: | 2023-06-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Harnessing Aromatic-Histidine Interactions through Synergistic Backbone Extension and Side Chain Modification.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8D51
 
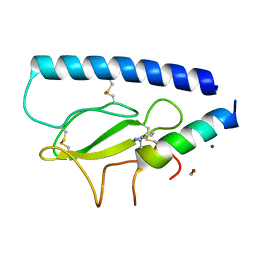 | | Parathyroid hormone 1 receptor extracellular domain complexed with a peptide ligand containing beta-3-homotryptophan | | Descriptor: | 1,2-ETHANEDIOL, PTHrP[1-36], Parathyroid hormone/parathyroid hormone-related peptide receptor, ... | | Authors: | Yu, Z, Kreitler, D.F, Gellman, S.H. | | Deposit date: | 2022-06-03 | | Release date: | 2023-06-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Harnessing Aromatic-Histidine Interactions through Synergistic Backbone Extension and Side Chain Modification.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
4MGP
 
 | | Structure of racemic Ala-(8,13,18) Magainin 2 | | Descriptor: | Magainin 2 Derivative | | Authors: | Hayouka, Z, Mortenson, D.E, Kreitler, D.F, Weisblum, B, Gellman, S.H, Forest, K.T. | | Deposit date: | 2013-08-28 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Evidence for Phenylalanine Zipper-Mediated Dimerization in the X-ray Crystal Structure of a Magainin 2 Analogue.
J.Am.Chem.Soc., 135, 2013
|
|
8FMF
 
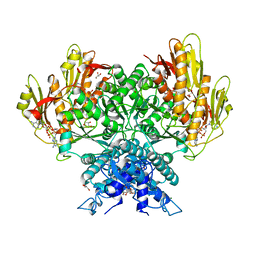 | | Structure of CBASS Cap5 from Pseudomonas syringae as an activated tetramer with the cyclic dinucleotide 3'2'-c-diAMP ligand (1 tetramer in the AU) | | Descriptor: | Cyclic (adenosine-(2'-5')-monophosphate-adenosine-(3'-5')-monophosphate, GLYCEROL, HEXAETHYLENE GLYCOL, ... | | Authors: | Rechkoblit, O, Kreitler, D.F, Aggarwal, A.K. | | Deposit date: | 2022-12-23 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Activation of CBASS Cap5 endonuclease immune effector by cyclic nucleotides.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8FM1
 
 | |
8FMG
 
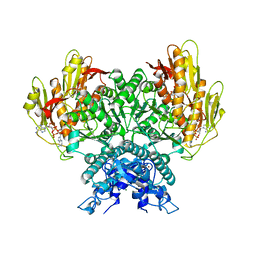 | | Structure of CBASS Cap5 from Pseudomonas syringae as an activated tetramer with the cyclic dinucleotide 3'2'-c-diAMP ligand (3 tetramers in the AU) | | Descriptor: | Cyclic (adenosine-(2'-5')-monophosphate-adenosine-(3'-5')-monophosphate, MAGNESIUM ION, SAVED domain-containing protein, ... | | Authors: | Rechkoblit, O, Kreitler, D.F, Aggarwal, A.K. | | Deposit date: | 2022-12-23 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Activation of CBASS Cap5 endonuclease immune effector by cyclic nucleotides.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8FMH
 
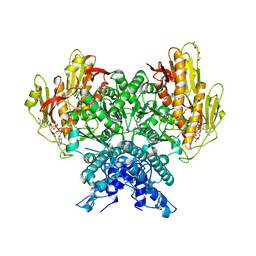 | | Structure of CBASS Cap5 from Pseudomonas syringae as an activated tetramer with the cyclic dinucleotide 3'2'-c-dGAMP ligand (2 tetramers in the AU) | | Descriptor: | 3'2'-cGAMP, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Rechkoblit, O, Kreitler, D.F, Aggarwal, A.K. | | Deposit date: | 2022-12-23 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Activation of CBASS Cap5 endonuclease immune effector by cyclic nucleotides.
Nat.Struct.Mol.Biol., 31, 2024
|
|
6NTX
 
 | |
6NIV
 
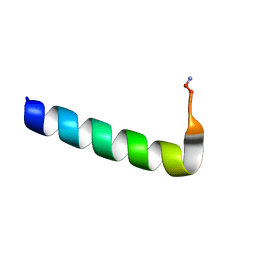 | | Racemic Phenol-Soluble Modulin Alpha 3 Peptide | | Descriptor: | Phenol-soluble modulin PSM-alpha-3 | | Authors: | Yao, Z, Cary, B.P, Bingman, C.A, Wang, C, Kreitler, D.F, Satyshur, K.A, Forest, K.T, Gellman, S.H. | | Deposit date: | 2018-12-31 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Use of a Stereochemical Strategy To Probe the Mechanism of Phenol-Soluble Modulin alpha 3 Toxicity.
J.Am.Chem.Soc., 141, 2019
|
|
6NRO
 
 | | Human parainfluenza virus type 3 fusion protein N-terminal heptad repeat domain+VIQKI | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, Human parainfluenza virus type 3 fusion glycoprotein C-terminal heptad repeat domain, ... | | Authors: | Outlaw, V.K, Kreitler, D.F, Gellman, S.H. | | Deposit date: | 2019-01-23 | | Release date: | 2019-07-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Dual Inhibition of Human Parainfluenza Type 3 and Respiratory Syncytial Virus Infectivity with a Single Agent.
J.Am.Chem.Soc., 141, 2019
|
|
6NYX
 
 | |
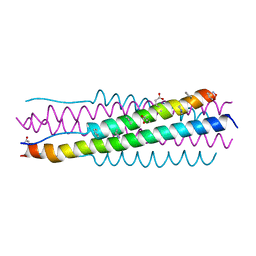
6VJO
 
 | |
7KU0
 
 | | Data clustering and dynamics of chymotrypsinogen cluster 138 (yellow) structure | | Descriptor: | Chymotrypsinogen A, SULFATE ION | | Authors: | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7KU2
 
 | | Data clustering and dynamics of chymotrypsinogen clulster 140 (structure) | | Descriptor: | Chymotrypsinogen A, SULFATE ION | | Authors: | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.185 Å) | | Cite: | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7KU3
 
 | | Data clustering and dynamics of chymotrypsinogen cluster 141 (cyan) structure | | Descriptor: | Chymotrypsinogen A, SULFATE ION | | Authors: | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7KU1
 
 | | Data clustering and dynamics of chymotrypsinogen cluster 139 (green) structure | | Descriptor: | Chymotrypsinogen A, SULFATE ION | | Authors: | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7KTZ
 
 | | Data clustering and dynamics of chymotrypsinogen cluster 131 (purple) structure | | Descriptor: | Chymotrypsinogen A, SULFATE ION | | Authors: | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7KTY
 
 | | Data clustering and dynamics of chymotrypsinogen average structure | | Descriptor: | Chymotrypsinogen A, SULFATE ION | | Authors: | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Shi, W, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7K40
 
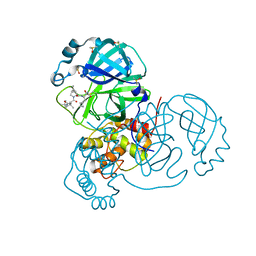 | | Crystal Structure of SARS-CoV-2 Main Protease (3CLpro/Mpro) in Complex with Covalent Inhibitor Boceprevir at 1.35 A Resolution | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, boceprevir (bound form) | | Authors: | Kumaran, D, Andi, B, Kreitler, D.F, Soares, A.S, Shi, W, Jakoncic, J, Fuchs, M.R, Keereetaweep, J, Shanklin, J, McSweeney, S. | | Deposit date: | 2020-09-14 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Hepatitis C virus NS3/4A inhibitors and other drug-like compounds as covalent binders of SARS-CoV-2 main protease.
Sci Rep, 12, 2022
|
|
7JYC
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (3CLpro/Mpro) in Complex with Covalent Inhibitor Narlaprevir | | Descriptor: | (1R,2S,5S)-3-[N-({1-[(tert-butylsulfonyl)methyl]cyclohexyl}carbamoyl)-3-methyl-L-valyl]-N-{(1S)-1-[(1R)-2-(cyclopropylamino)-1-hydroxy-2-oxoethyl]pentyl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE, ... | | Authors: | Andi, B, Kumaran, D, Kreitler, D.F, Soares, A.S, Shi, W, Jakoncic, J, Fuchs, M.R, Keereetaweep, J, Shanklin, J, McSweeney, S. | | Deposit date: | 2020-08-30 | | Release date: | 2020-09-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Hepatitis C virus NS3/4A inhibitors and other drug-like compounds as covalent binders of SARS-CoV-2 main protease.
Sci Rep, 12, 2022
|
|
7MNG
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (3CLpro/Mpro) in Complex with Covalent Inhibitor VBY-825 (Partial Occupancy) | | Descriptor: | (2R,3S)-N-cyclopropyl-3-{[(2R)-3-(cyclopropylmethanesulfonyl)-2-{[(1S)-2,2,2-trifluoro-1-(4-fluorophenyl)ethyl]amino}propanoyl]amino}-2-hydroxypentanamide (non-preferred name), 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Andi, B, Kumaran, D, Soares, A.S, Kreitler, D.F, Shi, W, Jakoncic, J, Fuchs, M.R, Keereetaweep, J, Shanklin, J, McSweeney, S. | | Deposit date: | 2021-04-30 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Hepatitis C virus NS3/4A inhibitors and other drug-like compounds as covalent binders of SARS-CoV-2 main protease.
Sci Rep, 12, 2022
|
|
7MRR
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (3CLpro/Mpro) in Complex with Covalent Inhibitor Leupeptin | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, LEUPEPTIN | | Authors: | Andi, B, Kumaran, D, Soares, A.S, Kreitler, D.F, Shi, W, Jakoncic, J, Fuchs, M.R, Keereetaweep, J, Shanklin, J, McSweeney, S. | | Deposit date: | 2021-05-08 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Hepatitis C virus NS3/4A inhibitors and other drug-like compounds as covalent binders of SARS-CoV-2 main protease.
Sci Rep, 12, 2022
|
|
7K3T
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (3CLpro/Mpro) at 1.2 A Resolution and a Possible Capture of Zinc Binding Intermediate | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | Authors: | Andi, B, Kumaran, D, Kreitler, D.F, Soares, A.S, Shi, W, Jakoncic, J, Fuchs, M.R, Keereetaweep, J, Shanklin, J, McSweeney, S. | | Deposit date: | 2020-09-13 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Hepatitis C virus NS3/4A inhibitors and other drug-like compounds as covalent binders of SARS-CoV-2 main protease.
Sci Rep, 12, 2022
|
|
4GWT
 
 | | Structure of racemic Pin1 WW domain cocrystallized with DL-malic acid | | Descriptor: | (2S)-2-hydroxybutanedioic acid, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Mortenson, D.E, Yun, H.G, Gellman, S.H, Forest, K.T. | | Deposit date: | 2012-09-03 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Evidence for small-molecule-mediated loop stabilization in the structure of the isolated Pin1 WW domain.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
