4YLE
 
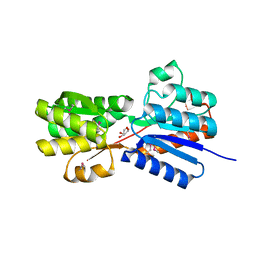 | | Crystal structure of an ABC transpoter solute binding protein (IPR025997) from Burkholderia multivorans (Bmul_1631, Target EFI-511115) with an unknown ligand modelled as alpha-D-erythrofuranose | | Descriptor: | Periplasmic binding protein/LacI transcriptional regulator, UNKNOWN LIGAND | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-03-05 | | Release date: | 2015-04-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of an ABC transporter solute binding protein (IPR025997) from Burkholderia multivorans (Bmul_1631, Target EFI-511115) with an unknown ligand modelled as alpha-D-erythrofuranose
To be published
|
|
4YQZ
 
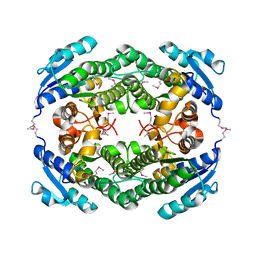 | | Crystal Structure of a putative oxidoreductase from Thermus Thermophilus HB27 (TT_P0034, TARGET EFI-513932) in its APO form | | Descriptor: | CHLORIDE ION, Putative oxidoreductase | | Authors: | Yadava, U, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-03-13 | | Release date: | 2015-03-25 | | Method: | X-RAY DIFFRACTION (1.807 Å) | | Cite: | Crystal Structure of a putative oxidoreductase from Thermus Thermophilus HB27 (TT_P0034, TARGET EFI-513932) in its APO form
To be published
|
|
4YWH
 
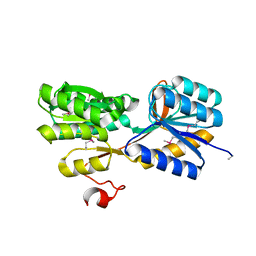 | | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM ACTINOBACILLUS SUCCINOGENES 130Z (Asuc_0499, TARGET EFI-511068) WITH BOUND D-XYLOSE | | Descriptor: | ABC TRANSPORTER SOLUTE BINDING PROTEIN, beta-D-xylopyranose | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-03-20 | | Release date: | 2015-04-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM ACTINOBACILLUS SUCCINOGENES 130Z (Asuc_0499, TARGET EFI-511068) WITH BOUND D-XYLOSE
To be published
|
|
4YIC
 
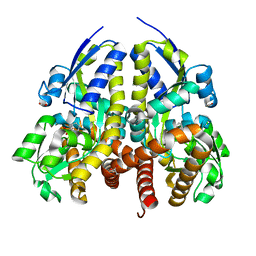 | | CRYSTAL STRUCTURE OF A TRAP TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM BORDETELLA BRONCHISEPTICA RB50 (BB0280, TARGET EFI-500035) WITH BOUND PICOLINIC ACID | | Descriptor: | ACETATE ION, CALCIUM ION, IMIDAZOLE, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-03-01 | | Release date: | 2015-04-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | CRYSTAL STRUCTURE OF A TRAP TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM BORDETELLA BRONCHISEPTICA RB50 (BB0280, TARGET EFI-500035) WITH BOUND PICOLINIC ACID
To be published
|
|
4YV7
 
 | | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM MYCOBACTERIUM SMEGMATIS (MSMEI_3018, TARGET EFI-511327) WITH BOUND GLYCEROL | | Descriptor: | GLYCEROL, Periplasmic binding protein/LacI transcriptional regulator | | Authors: | Vetting, M.W, Patskovsky, Y, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-03-19 | | Release date: | 2015-04-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM MYCOBACTERIUM SMEGMATIS (MSMEI_3018, TARGET EFI-511327) WITH BOUND GLYCEROL
To be published
|
|
4Z0N
 
 | | Crystal Structure of a Periplasmic Solute binding protein (IPR025997) from Streptobacillus moniliformis DSM-12112 (Smon_0317, TARGET EFI-511281) with bound D-Galactose | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Yadava, U, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-03-26 | | Release date: | 2015-04-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Crystal Structure of a Periplasmic Solute binding protein (IPR025997) from Streptobacillus moniliformis DSM-12112 (Smon_0317, TARGET EFI-511281) with bound D-Galactose
To be published
|
|
5BRA
 
 | | Crystal Structure of a putative Periplasmic Solute binding protein (IPR025997) from Ochrobactrum Anthropi ATCC49188 (Oant_2843, TARGET EFI-511085) | | Descriptor: | Putative periplasmic binding protein with substrate ribose | | Authors: | Yadava, U, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-05-30 | | Release date: | 2015-06-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.971 Å) | | Cite: | Crystal Structure of a putative Periplasmic Solute binding protein (IPR025997) from Ochrobactrum Anthropi ATCC49188(Oant_2843, TARGET EFI-511085)
To be published
|
|
3DTD
 
 | | Crystal structure of invasion associated protein b from bartonella henselae | | Descriptor: | GLYCEROL, Invasion-associated protein B | | Authors: | Patskovsky, Y, Ozyurt, S, Freeman, J, Slocombe, A, Groshong, C, Koss, J, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-07-14 | | Release date: | 2008-09-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of Invasion Associated Protein B from Bartonella Henselae.
To be Published
|
|
3DBI
 
 | | CRYSTAL STRUCTURE OF SUGAR-BINDING TRANSCRIPTIONAL REGULATOR (LACI FAMILY) FROM ESCHERICHIA COLI COMPLEXED WITH PHOSPHATE | | Descriptor: | GLYCEROL, PHOSPHATE ION, SUGAR-BINDING TRANSCRIPTIONAL REGULATOR, ... | | Authors: | Patskovsky, Y, Ozyurt, S, Freeman, J, Wu, B, Maletic, M, Koss, J, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-06-01 | | Release date: | 2008-07-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structure of Sugar-Binding Transcriptional Regulator (LacI Family) from Escherichia Coli Complexed with Phosphate.
To be Published
|
|
3C19
 
 | | Crystal structure of protein MK0293 from Methanopyrus kandleri AV19 | | Descriptor: | GLYCEROL, PHOSPHATE ION, Uncharacterized protein MK0293 | | Authors: | Patskovsky, Y, Romero, R, Bonanno, J.B, Malashkevich, V, Dickey, M, Chang, S, Koss, J, Bain, K, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-01-22 | | Release date: | 2008-02-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of protein MK0293 from Methanopyrus kandleri AV19.
To be Published
|
|
3BMA
 
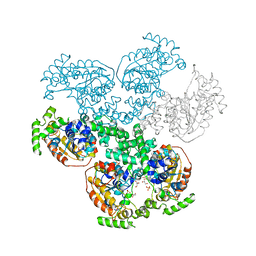 | | Crystal structure of D-alanyl-lipoteichoic acid synthetase from Streptococcus pneumoniae R6 | | Descriptor: | D-alanyl-lipoteichoic acid synthetase, GLYCEROL, SULFATE ION | | Authors: | Patskovsky, Y, Sridhar, V, Bonanno, J.B, Smith, D, Rutter, M, Iizuka, M, Koss, J, Bain, K, Gheyi, T, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-12-12 | | Release date: | 2007-12-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Crystal Structure of probable D-Alanyl-Lipoteichoic Acid Synthetase from Streptococcus pneumoniae.
To be Published
|
|
3BQ9
 
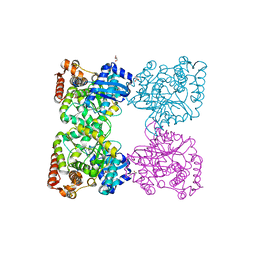 | | Crystal structure of predicted nucleotide-binding protein from Idiomarina baltica OS145 | | Descriptor: | GLYCEROL, Predicted Rossmann fold nucleotide-binding domain-containing protein, SULFATE ION | | Authors: | Patskovsky, Y, Toro, R, Meyer, A.J, Dickey, M, Eberle, M, Koss, J, Groshong, C, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-12-19 | | Release date: | 2008-01-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Predicted Nucleotide-Binding Protein from Idiomarina baltica.
To be Published
|
|
3CG0
 
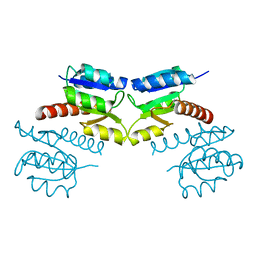 | | Crystal structure of signal receiver domain of modulated diguanylate cyclase from Desulfovibrio desulfuricans G20, an example of alternate folding | | Descriptor: | Response regulator receiver modulated diguanylate cyclase with PAS/PAC sensor | | Authors: | Patskovsky, Y, Bonanno, J.B, Romero, R, Gilmore, M, Chang, S, Groshong, C, Koss, J, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-03-04 | | Release date: | 2008-03-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of Signal Receiver Domain of Modulated Diguanylate Cyclase from Desulfovibrio desulfuricans.
To be Published
|
|
3DJC
 
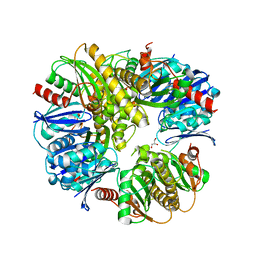 | | CRYSTAL STRUCTURE OF PANTOTHENATE KINASE FROM LEGIONELLA PNEUMOPHILA | | Descriptor: | GLYCEROL, Type III pantothenate kinase | | Authors: | Patskovsky, Y, Bonanno, J.B, Romero, R, Dickey, M, Logan, C, Wasserman, S, Maletic, M, Koss, J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-06-23 | | Release date: | 2008-07-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Pantothenate Kinase from Legionella Pneumophila
To be Published
|
|
3ED4
 
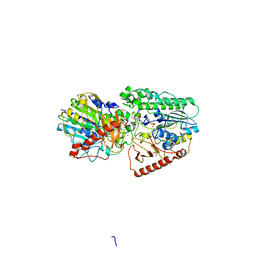 | | Crystal structure of putative arylsulfatase from escherichia coli | | Descriptor: | ARYLSULFATASE, GLYCEROL, SODIUM ION, ... | | Authors: | Patskovsky, Y, Ozyurt, S, Gilmore, M, Chang, S, Bain, K, Wasserman, S, Koss, J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-09-02 | | Release date: | 2008-09-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Arylsulfatase from Escherichia Coli
To be Published
|
|
3FRN
 
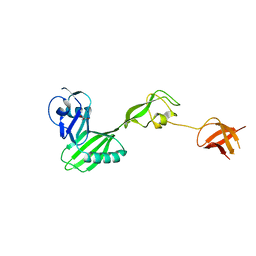 | | CRYSTAL STRUCTURE OF flagellar protein FlgA FROM Thermotoga maritima MSB8 | | Descriptor: | Flagellar protein FlgA, GLYCEROL | | Authors: | Patskovsky, Y, Bonanno, J.B, Romero, R, Gilmore, M, Hu, S, Bain, K, Koss, J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-01-08 | | Release date: | 2009-02-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | CRYSTAL STRUCTURE OF flagellar protein FlgA FROM Thermotoga maritima
To be Published
|
|
2FH7
 
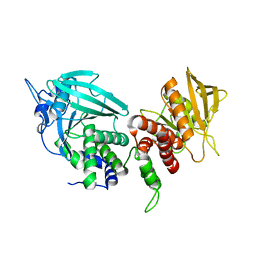 | | Crystal structure of the phosphatase domains of human PTP SIGMA | | Descriptor: | Receptor-type tyrosine-protein phosphatase S | | Authors: | Alvarado, J, Udupi, R, Smith, D, Koss, J, Wasserman, S.R, Ozyurt, S, Atwell, S, Powell, A, Kearins, M.C, Rooney, I, Maletic, M, Bain, K.T, Freeman, J.C, Russell, M, Thompson, D.A, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-12-23 | | Release date: | 2006-01-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural genomics of protein phosphatases.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
2I1Y
 
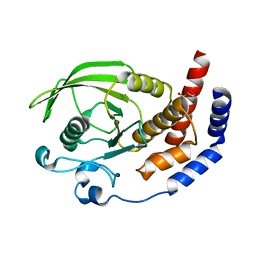 | | Crystal structure of the phosphatase domain of human PTP IA-2 | | Descriptor: | GLYCEROL, Receptor-type tyrosine-protein phosphatase | | Authors: | Faber-Barata, J, Patskovsky, Y, Alvarado, J, Smith, D, Koss, J, Wasserman, S.R, Ozyurt, S, Atwell, S, Powell, A, Kearins, M.C, Maletic, M, Rooney, I, Bain, K.T, Freeman, M, Russell, J.C, Thompson, D.A, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-08-15 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structural genomics of protein phosphatases
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
6MR9
 
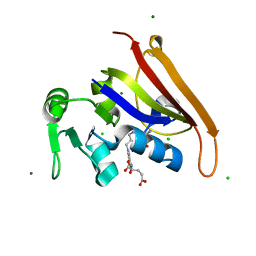 | | E. coli DHFR complex with a reaction intermediate | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, CHLORIDE ION, Dihydrofolate reductase, ... | | Authors: | Cao, H, Rodrigues, J, Benach, J, Frommelt, A, Morisco, L, Koss, J, Shakhnovich, E, Skolnick, J. | | Deposit date: | 2018-10-12 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Time-resolved x-ray crystallography capture of a slow reaction tetrahydrofolate intermediate.
Struct Dyn., 6, 2019
|
|
6MT8
 
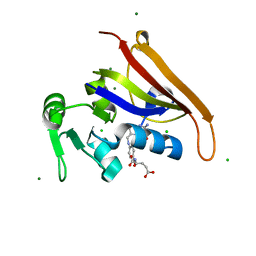 | | E. coli DHFR complex modeled with two ligand states | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, CHLORIDE ION, DIHYDROFOLIC ACID, ... | | Authors: | Cao, H, Rodrigues, J, Benach, J, Frommelt, A, Morisco, L, Koss, J, Shakhnovich, E, Skolnick, J. | | Deposit date: | 2018-10-19 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Time-resolved x-ray crystallography capture of a slow reaction tetrahydrofolate intermediate.
Struct Dyn., 6, 2019
|
|
6MTH
 
 | | E. coli DHFR complex modeled with three ligand states | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, CHLORIDE ION, DIHYDROFOLIC ACID, ... | | Authors: | Cao, H, Rodrigues, J, Benach, J, Frommelt, A, Morisco, L, Koss, J, Shakhnovich, E, Skolnick, J. | | Deposit date: | 2018-10-19 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Time-resolved x-ray crystallography capture of a slow reaction tetrahydrofolate intermediate.
Struct Dyn., 6, 2019
|
|
3NEK
 
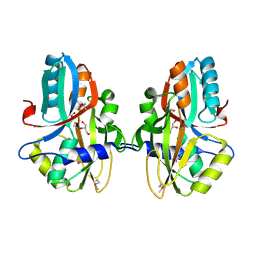 | | Crystal structure of a nitrogen repressor-like protein MJ0159 from Methanococcus jannaschii | | Descriptor: | GLYCEROL, nitrogen repressor-like protein MJ0159 | | Authors: | Bonanno, J.B, Patskovsky, Y, Malashkevich, V, Ozyurt, S, Dickey, M, Wu, B, Maletic, M, Rodgers, L, Koss, J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-06-09 | | Release date: | 2010-06-23 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural underpinnings of nitrogen regulation by the prototypical nitrogen-responsive transcriptional factor NrpR.
Structure, 18, 2010
|
|
6CW7
 
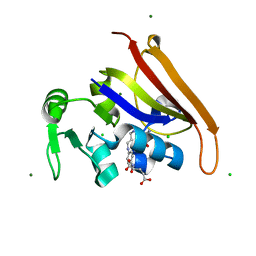 | | E. coli DHFR product complex with (6S)-5,6,7,8-TETRAHYDROFOLATE | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, CHLORIDE ION, Dihydrofolate reductase, ... | | Authors: | Cao, H, Rodrigues, J, Benach, J, Frommelt, A, Morisco, L, Koss, J, Shakhnovich, E, Skolnick, J. | | Deposit date: | 2018-03-30 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | The crystal structure of a tetrahydrofolate-bound dihydrofolate reductase reveals the origin of slow product release.
Commun Biol, 1, 2018
|
|
6CYV
 
 | | E. coli DHFR ternary complex with NADP and dihydrofolate | | Descriptor: | DIHYDROFOLIC ACID, Dihydrofolate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cao, H, Rodrigues, J, Benach, J, Frommelt, A, Morisco, L, Koss, J, Shakhnovich, E, Skolnick, J. | | Deposit date: | 2018-04-06 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The crystal structure of a tetrahydrofolate-bound dihydrofolate reductase reveals the origin of slow product release.
Commun Biol, 1, 2018
|
|
6CQA
 
 | | E. coli DHFR complex with inhibitor AMPQD | | Descriptor: | 7-[(3-aminophenyl)methyl]-7H-pyrrolo[3,2-f]quinazoline-1,3-diamine, Dihydrofolate reductase, SULFATE ION | | Authors: | Cao, H, Rodrigues, J, Benach, J, Wasserman, S, Morisco, L, Koss, J, Shakhnovich, E, Skolnick, J. | | Deposit date: | 2018-03-14 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of a tetrahydrofolate-bound dihydrofolate reductase reveals the origin of slow product release.
Commun Biol, 1, 2018
|
|
