2EDC
 
 | |
2EDA
 
 | |
7JKM
 
 | |
1LKK
 
 | |
1LKL
 
 | |
2KLJ
 
 | | Solution Structure of gammaD-Crystallin with RDC and SAXS | | Descriptor: | Gamma-crystallin D | | Authors: | Wang, J, Zuo, X, Yu, P, Byeon, I, Jung, J, Gronenborn, A.M, Wang, Y. | | Deposit date: | 2009-07-06 | | Release date: | 2009-10-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Determination of multicomponent protein structures in solution using global orientation and shape restraints.
J.Am.Chem.Soc., 131, 2009
|
|
1B6G
 
 | | HALOALKANE DEHALOGENASE AT PH 5.0 CONTAINING CHLORIDE | | Descriptor: | CHLORIDE ION, GLYCEROL, HALOALKANE DEHALOGENASE, ... | | Authors: | Ridder, I.S, Rozeboom, H.J, Dijkstra, B.W. | | Deposit date: | 1999-01-14 | | Release date: | 1999-07-13 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Haloalkane dehalogenase from Xanthobacter autotrophicus GJ10 refined at 1.15 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1AQ6
 
 | | STRUCTURE OF L-2-HALOACID DEHALOGENASE FROM XANTHOBACTER AUTOTROPHICUS | | Descriptor: | FORMIC ACID, L-2-HALOACID DEHALOGENASE | | Authors: | Ridder, I.S, Rozeboom, H.J, Kalk, K.H, Dijkstra, B.W. | | Deposit date: | 1997-08-07 | | Release date: | 1998-01-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Three-dimensional structure of L-2-haloacid dehalogenase from Xanthobacter autotrophicus GJ10 complexed with the substrate-analogue formate.
J.Biol.Chem., 272, 1997
|
|
8UWC
 
 | | Site-one protease without SPRING | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Membrane-bound transcription factor site-1 protease | | Authors: | Kober, D.L. | | Deposit date: | 2023-11-06 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.27 Å) | | Cite: | SPRING licenses S1P-mediated cleavage of SREBP2 by displacing an inhibitory pro-domain.
Nat Commun, 15, 2024
|
|
7O7K
 
 | | Crystal structure of the human DYRK1A kinase domain bound to abemaciclib | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kaltheuner, I.H, Anand, K, Geyer, M. | | Deposit date: | 2021-04-13 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Abemaciclib is a potent inhibitor of DYRK1A and HIP kinases involved in transcriptional regulation.
Nat Commun, 12, 2021
|
|
7O7I
 
 | |
7O7J
 
 | | Crystal structure of the human HIPK3 kinase domain bound to abemaciclib | | Descriptor: | Homeodomain-interacting protein kinase 3, N-{5-[(4-ethylpiperazin-1-yl)methyl]pyridin-2-yl}-5-fluoro-4-[4-fluoro-2-methyl-1-(propan-2-yl)-1H-benzimidazol-6-yl]py rimidin-2-amine | | Authors: | Kaltheuner, I.H, Anand, K, Geyer, M. | | Deposit date: | 2021-04-13 | | Release date: | 2021-11-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Abemaciclib is a potent inhibitor of DYRK1A and HIP kinases involved in transcriptional regulation.
Nat Commun, 12, 2021
|
|
7AC0
 
 | | Epoxide hydrolase CorEH without ligand | | Descriptor: | Soluble epoxide hydrolase | | Authors: | Palm, G.J, Lammers, M, Berndt, L. | | Deposit date: | 2020-09-09 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.177 Å) | | Cite: | Promiscuous Dehalogenase Activity of the Epoxide Hydrolase CorEH from Corynebacterium sp. C12
Acs Catalysis, 11, 2021
|
|
8UW8
 
 | | Site-one protease and SPRING | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Membrane-bound transcription factor site-1 protease, ... | | Authors: | Kober, D.L. | | Deposit date: | 2023-11-06 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | SPRING licenses S1P-mediated cleavage of SREBP2 by displacing an inhibitory pro-domain.
Nat Commun, 15, 2024
|
|
1EDD
 
 | |
1EDB
 
 | |
3K2M
 
 | |
7PZC
 
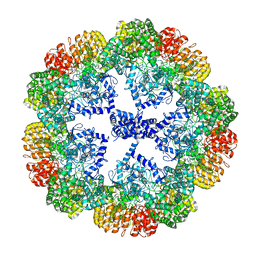 | | Cryo-EM structure of the NLRP3 decamer bound to the inhibitor CRID3 | | Descriptor: | 1-(1,2,3,5,6,7-hexahydro-s-indacen-4-yl)-3-[4-(2-oxidanylpropan-2-yl)furan-2-yl]sulfonyl-urea, ADENOSINE-5'-DIPHOSPHATE, NACHT, ... | | Authors: | Hochheiser, I.V, Pilsl, M, Hagelueken, G, Engel, C, Geyer, M. | | Deposit date: | 2021-10-12 | | Release date: | 2022-01-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the NLRP3 decamer bound to the cytokine release inhibitor CRID3.
Nature, 604, 2022
|
|
7QE5
 
 | |
7QDA
 
 | | Crystal structure of CalpL | | Descriptor: | CalpL, SULFATE ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Schneberger, N, Hagelueken, G. | | Deposit date: | 2021-11-26 | | Release date: | 2022-11-16 | | Last modified: | 2023-02-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Antiviral signalling by a cyclic nucleotide activated CRISPR protease.
Nature, 614, 2023
|
|
8P81
 
 | | Crystal structure of human Cdk12/Cyclin K in complex with inhibitor SR-4835 | | Descriptor: | Cyclin-K, Cyclin-dependent kinase 12, ~{N}-[[5,6-bis(chloranyl)-1~{H}-benzimidazol-2-yl]methyl]-9-(1-methylpyrazol-4-yl)-2-morpholin-4-yl-purin-6-amine | | Authors: | Anand, K, Schmitz, M, Geyer, M. | | Deposit date: | 2023-05-31 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | The reversible inhibitor SR-4835 binds Cdk12/cyclin K in a noncanonical G-loop conformation.
J.Biol.Chem., 300, 2023
|
|
2HAD
 
 | |
7Z1X
 
 | |
8B0R
 
 | | Structure of the CalpL/cA4 complex | | Descriptor: | Cyclic tetraadenosine monophosphate (cA4), SMODS-associated and fused to various effectors domain-containing protein, SULFATE ION, ... | | Authors: | Schneberger, N, Hagelueken, G. | | Deposit date: | 2022-09-08 | | Release date: | 2022-11-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Antiviral signalling by a cyclic nucleotide activated CRISPR protease.
Nature, 614, 2023
|
|
8B0U
 
 | | Structure of the CalpL/T10 complex | | Descriptor: | CalpT10, GLYCEROL, SAVED domain-containing protein, ... | | Authors: | Schneberger, N, Hagelueken, G. | | Deposit date: | 2022-09-08 | | Release date: | 2022-11-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Antiviral signalling by a cyclic nucleotide activated CRISPR protease.
Nature, 614, 2023
|
|
