9EEG
 
 | | Room-temperature X-ray structure of HIV-1 protease in complex with GRL-11124A inhibitor | | Descriptor: | (3S,3aS,4R,7R,8aS)-3-methylhexahydro-1H,3H-3a,7-epoxycyclohepta[c]furan-4-yl {(2S,3R)-3-hydroxy-4-[(4-methoxybenzene-1-sulfonyl)(2-methylpropyl)amino]-1-phenylbutan-2-yl}carbamate, CHLORIDE ION, Protease | | Authors: | Kovalevsky, A, Gerlits, O, Ghosh, A. | | Deposit date: | 2024-11-19 | | Release date: | 2025-02-05 | | Last modified: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Potent HIV‐1 protease inhibitors containing oxabicyclo octanol-derived P2-ligands: Design, synthesis, and X‐ray structural studies of inhibitor-HIV-1 protease complexes.
Bioorg.Med.Chem.Lett., 120, 2025
|
|
7UHC
 
 | |
8FUJ
 
 | | HIV-1 wild type protease with GRL-03419A, with N-(2,5-dimethylphenyl)-4-(pyridin-3-yl)pyrimidin-2-amine as P2-P3 group and 3,5-difluorophenylmethyl as the P1 group | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Ghosh, A.K, Weber, I.T. | | Deposit date: | 2023-01-17 | | Release date: | 2023-05-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Exploration of imatinib and nilotinib-derived templates as the P2-Ligand for HIV-1 protease inhibitors: Design, synthesis, protein X-ray structural studies, and biological evaluation.
Eur.J.Med.Chem., 255, 2023
|
|
8FUI
 
 | | HIV-1 wild type protease with GRL-02519A, with N-(2,5-dimethylphenyl)-4-(pyridin-3-yl)pyrimidin-2-amine as P2-P3 group | | Descriptor: | ACETATE ION, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Wang, Y.-F, Wong-Sam, A.E, Ghosh, A.K, Weber, I.T. | | Deposit date: | 2023-01-17 | | Release date: | 2023-05-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Exploration of imatinib and nilotinib-derived templates as the P2-Ligand for HIV-1 protease inhibitors: Design, synthesis, protein X-ray structural studies, and biological evaluation.
Eur.J.Med.Chem., 255, 2023
|
|
6HSV
 
 | | Engineered higher-order assembly of Cholera Toxin B subunits via the addition of C-terminal parallel coiled-coiled domains | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, PHOSPHATE ION, ... | | Authors: | Pearson, A.R, Turnbull, W.B, Ross, J.F, Trinh, C.H, Webb, M.E. | | Deposit date: | 2018-10-01 | | Release date: | 2019-03-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Directed Assembly of Homopentameric Cholera Toxin B-Subunit Proteins into Higher-Order Structures Using Coiled-Coil Appendages.
J.Am.Chem.Soc., 141, 2019
|
|
7LBE
 
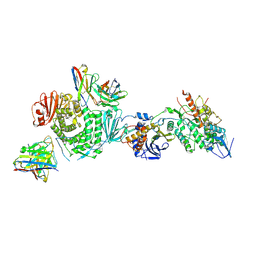 | | CryoEM structure of the HCMV Trimer gHgLgO in complex with neutralizing fabs 13H11 and MSL-109 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein H, ... | | Authors: | Kschonsak, M, Rouge, L, Arthur, C.P, Hoangdung, H, Patel, N, Kim, I, Johnson, M, Kraft, E, Rohou, A.L, Gill, A, Martinez-Martin, N, Payandeh, J, Ciferri, C. | | Deposit date: | 2021-01-07 | | Release date: | 2021-03-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of HCMV Trimer reveal the basis for receptor recognition and cell entry.
Cell, 184, 2021
|
|
7LBG
 
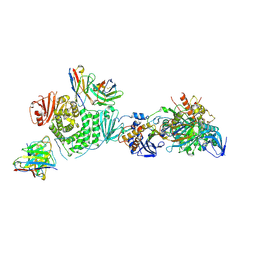 | | CryoEM structure of the HCMV Trimer gHgLgO in complex with human Transforming growth factor beta receptor type 3 and neutralizing fabs 13H11 and MSL-109 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein H, ... | | Authors: | Kschonsak, M, Rouge, L, Arthur, C.P, Hoangdung, H, Patel, N, Kim, I, Johnson, M, Kraft, E, Rohou, A.L, Gill, A, Martinez-Martin, N, Payandeh, J, Ciferri, C. | | Deposit date: | 2021-01-07 | | Release date: | 2021-03-10 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structures of HCMV Trimer reveal the basis for receptor recognition and cell entry.
Cell, 184, 2021
|
|
7LBF
 
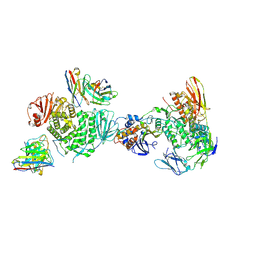 | | CryoEM structure of the HCMV Trimer gHgLgO in complex with human Platelet-derived growth factor receptor alpha and neutralizing fabs 13H11 and MSL-109 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein H, ... | | Authors: | Kschonsak, M, Rouge, L, Arthur, C.P, Hoangdung, H, Patel, N, Kim, I, Johnson, M, Kraft, E, Rohou, A.L, Gill, A, Martinez-Martin, N, Payandeh, J, Ciferri, C. | | Deposit date: | 2021-01-07 | | Release date: | 2021-03-10 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures of HCMV Trimer reveal the basis for receptor recognition and cell entry.
Cell, 184, 2021
|
|
9CNY
 
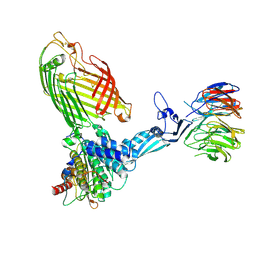 | | CryoEM structure of the APO-BAM complex in SMA nanodisc | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | Authors: | Sun, D, Tegunov, D, Payandeh, J. | | Deposit date: | 2024-07-15 | | Release date: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | The discovery and structural basis of two distinct state-dependent inhibitors of BamA.
Nat Commun, 15, 2024
|
|
9CNZ
 
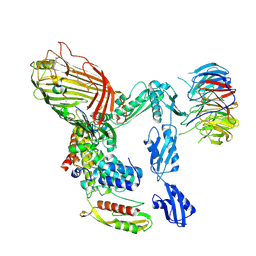 | | Structure of BAM complexed with PTB2 ligand in detergent | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | Authors: | Sun, D, Tegunov, D, Payandeh, J. | | Deposit date: | 2024-07-15 | | Release date: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The discovery and structural basis of two distinct state-dependent inhibitors of BamA.
Nat Commun, 15, 2024
|
|
9CNW
 
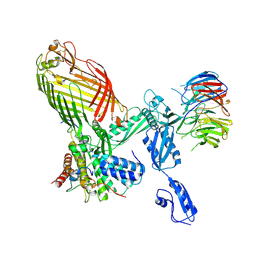 | | CryoEM structure of the APO-BAM complex in DDM detergent | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | Authors: | Sun, D, Tegunov, D, Payandeh, J. | | Deposit date: | 2024-07-15 | | Release date: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | The discovery and structural basis of two distinct state-dependent inhibitors of BamA.
Nat Commun, 15, 2024
|
|
9CO0
 
 | | CryoEM structure of BAM in complex with the PTB2 open-state inhibitor (in SMA nanodisc) | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | Authors: | Sun, D, Tegunov, D, Payandeh, J. | | Deposit date: | 2024-07-15 | | Release date: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The discovery and structural basis of two distinct state-dependent inhibitors of BamA.
Nat Commun, 15, 2024
|
|
9CNX
 
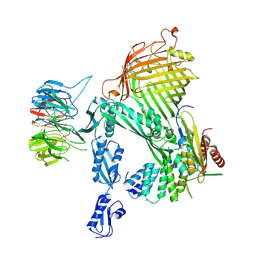 | | CryoEM structure of BAM in complex with the PTB1 closed-state inhibitor (in DDM detergent) | | Descriptor: | Circular peptide, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Sun, D, Tegunov, D, Payandeh, J. | | Deposit date: | 2024-07-15 | | Release date: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | The discovery and structural basis of two distinct state-dependent inhibitors of BamA.
Nat Commun, 15, 2024
|
|
9CO2
 
 | |
6PZ4
 
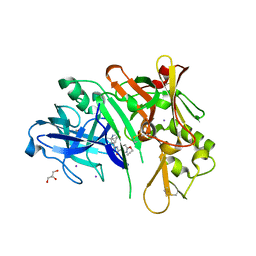 | | co-crystal structure of BACE with inhibitor AM-6494 | | Descriptor: | Beta-secretase 1, GLYCEROL, IODIDE ION, ... | | Authors: | Huang, X. | | Deposit date: | 2019-07-31 | | Release date: | 2019-10-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of AM-6494: A Potent and Orally Efficacious beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 (BACE1) Inhibitor with in Vivo Selectivity over BACE2.
J.Med.Chem., 63, 2020
|
|
5OW4
 
 | |
5OW3
 
 | | Crystal structure of a C-terminally truncated trimeric ectodomain of the Arabidopsis thaliana gamete fusion protein HAP2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, GLYCEROL, ... | | Authors: | Fedry, J, Legrand, P, Rey, F.A, Krey, T. | | Deposit date: | 2017-08-30 | | Release date: | 2018-08-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Evolutionary diversification of the HAP2 membrane insertion motifs to drive gamete fusion across eukaryotes.
PLoS Biol., 16, 2018
|
|
3IN4
 
 | | Bace1 with Compound 38 | | Descriptor: | (5S)-2-amino-5-(2,6-diethylpyridin-4-yl)-3-methyl-5-(3-pyrimidin-5-ylphenyl)-3,5-dihydro-4H-imidazol-4-one, Beta-secretase 1 | | Authors: | Olland, A.M. | | Deposit date: | 2009-08-11 | | Release date: | 2010-01-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Di-substituted pyridinyl aminohydantoins as potent and highly selective human beta-secretase (BACE1) inhibitors.
Bioorg.Med.Chem., 18, 2010
|
|
3IN3
 
 | | Bace1 with Compound 30 | | Descriptor: | (5S)-2-amino-3-methyl-5-pyridin-4-yl-5-(3-pyridin-3-ylphenyl)-3,5-dihydro-4H-imidazol-4-one, Beta-secretase 1 | | Authors: | Olland, A.M. | | Deposit date: | 2009-08-11 | | Release date: | 2010-01-19 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Di-substituted pyridinyl aminohydantoins as potent and highly selective human beta-secretase (BACE1) inhibitors.
Bioorg.Med.Chem., 18, 2010
|
|
6N3R
 
 | |
6N44
 
 | |
3RM8
 
 | | AMCase in complex with Compound 2 | | Descriptor: | 2-methyl-3-{[4-(pyridin-2-yl)piperazin-1-yl]methyl}-1H-indole, Acidic mammalian chitinase | | Authors: | Olland, A. | | Deposit date: | 2011-04-20 | | Release date: | 2011-08-24 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification and Characterization of Acidic Mammalian Chitinase Inhibitors
J.Med.Chem., 53, 2010
|
|
3S7L
 
 | | Pyrazolyl and Thienyl Aminohydantoins as Potent BACE1 Inhibitors | | Descriptor: | (5S)-2-amino-5-(1-ethyl-1H-pyrazol-4-yl)-3-methyl-5-[3-(pyrimidin-5-yl)phenyl]-3,5-dihydro-4H-imidazol-4-one, Beta-secretase 1 | | Authors: | Chopra, R, Olland, A, Svenson, K. | | Deposit date: | 2011-05-26 | | Release date: | 2011-08-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.162 Å) | | Cite: | New pyrazolyl and thienyl aminohydantoins as potent BACE1 inhibitors: Exploring the S2' region.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3IGB
 
 | | Bace-1 with Compound 3 | | Descriptor: | 8,8-diphenyl-2,3,4,8-tetrahydroimidazo[1,5-a]pyrimidin-6-amine, Beta-secretase 1 | | Authors: | Olland, A.M. | | Deposit date: | 2009-07-27 | | Release date: | 2009-11-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.238 Å) | | Cite: | Aminoimidazoles as potent and selective human beta-secretase (BACE1) inhibitors
J.Med.Chem., 52, 2009
|
|
3RME
 
 | | AMCase in complex with Compound 5 | | Descriptor: | Acidic mammalian chitinase, GLYCEROL, N-ethyl-2-(4-methylpiperazin-1-yl)pyridine-3-carboxamide | | Authors: | Olland, A. | | Deposit date: | 2011-04-20 | | Release date: | 2011-08-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification and Characterization of Acidic Mammalian Chitinase Inhibitors
J.Med.Chem., 53, 2010
|
|
