5U5H
 
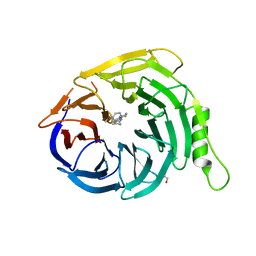 | | Crystal structure of EED in complex with 6-(2-fluoro-5-methoxybenzyl)-1-isopropyl-5,6,7,8-tetrahydroimidazo[1,5-a]pyridin-3-amine 6-(2-fluoro-5-methoxybenzyl)-1-isopropyl-5,6,7,8-tetrahydroimidazo[1,5-a]pyridin-3-amine | | Descriptor: | (6S)-6-[(2-fluoro-5-methoxyphenyl)methyl]-1-(propan-2-yl)-5,6,7,8-tetrahydroimidazo[1,5-a]pyridin-3-amine, GLYCEROL, Polycomb protein EED | | Authors: | Bussiere, D, Shu, W. | | Deposit date: | 2016-12-06 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Guided Design of EED Binders Allosterically Inhibiting the Epigenetic Polycomb Repressive Complex 2 (PRC2) Methyltransferase.
J. Med. Chem., 60, 2017
|
|
5U62
 
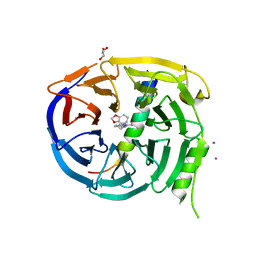 | | Crystal structure of EED in complex with H3K27Me3 peptide and 6-(benzo[d][1,3]dioxol-4-ylmethyl)-5,6,7,8-tetrahydroimidazo[1,5-a]pyridin-3-amine | | Descriptor: | (6S)-6-[(2H-1,3-benzodioxol-4-yl)methyl]-5,6,7,8-tetrahydroimidazo[1,5-a]pyridin-3-amine, GLYCEROL, Histone-lysine N-methyltransferase EZH2, ... | | Authors: | Bussiere, D, Shu, W. | | Deposit date: | 2016-12-07 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Guided Design of EED Binders Allosterically Inhibiting the Epigenetic Polycomb Repressive Complex 2 (PRC2) Methyltransferase.
J. Med. Chem., 60, 2017
|
|
7WFC
 
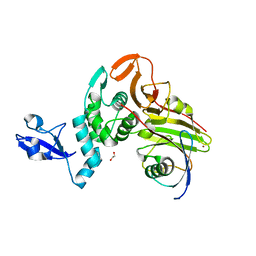 | | X-ray structure of HKU1-PLP2(Cys109Ser) catalytic mutant in complex with free ubiquitin | | Descriptor: | 1,2-ETHANEDIOL, 60S ribosomal protein L40, Papain-like protease, ... | | Authors: | Xiong, Y.X, Fu, Z.Y, Huang, H. | | Deposit date: | 2021-12-26 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The substrate selectivity of papain-like proteases from human-infecting coronaviruses correlates with innate immune suppression.
Sci.Signal., 16, 2023
|
|
6O8B
 
 | | Crystal structure of STING CTD in complex with TBK1 | | Descriptor: | N-(3-{[5-iodo-4-({3-[(thiophen-2-ylcarbonyl)amino]propyl}amino)pyrimidin-2-yl]amino}phenyl)pyrrolidine-1-carboxamide, Serine/threonine-protein kinase TBK1, Stimulator of interferon genes protein | | Authors: | Li, P, Zhao, B, Du, F. | | Deposit date: | 2019-03-09 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | A conserved PLPLRT/SD motif of STING mediates the recruitment and activation of TBK1.
Nature, 569, 2019
|
|
6O8C
 
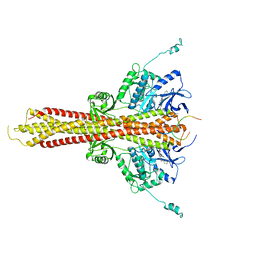 | | Crystal structure of STING CTT in complex with TBK1 | | Descriptor: | N-(3-{[5-iodo-4-({3-[(thiophen-2-ylcarbonyl)amino]propyl}amino)pyrimidin-2-yl]amino}phenyl)pyrrolidine-1-carboxamide, Serine/threonine-protein kinase TBK1, Stimulator of interferon genes protein | | Authors: | Li, P, Zhao, B, Du, F. | | Deposit date: | 2019-03-09 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | A conserved PLPLRT/SD motif of STING mediates the recruitment and activation of TBK1.
Nature, 569, 2019
|
|
8HI8
 
 | |
8HCI
 
 | |
8HI7
 
 | |
6LE3
 
 | |
7ASL
 
 | |
7ASH
 
 | |
6ZTY
 
 | |
6ZTS
 
 | |
6WPC
 
 | |
6WWB
 
 | | Crystal Structure of the second bromodomain of human BRD2 in complex with the compound 3b | | Descriptor: | 1,2-ETHANEDIOL, 2-((S)-4-(4-chlorophenyl)-2,3,9-trimethyl-6H-thieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-6-yl)-N-((1-(4-(2-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)acetamido)butyl)-1H-1,2,3-triazol-4-yl)methyl)acetamide, Bromodomain-containing protein 2 | | Authors: | White, S.W, Yun, M. | | Deposit date: | 2020-05-08 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Phenyl-Glutarimides: Alternative Cereblon Binders for the Design of PROTACs.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6K9N
 
 | | Rice_OTUB_like_catalytic domain | | Descriptor: | Ubiquitin thioesterase | | Authors: | Lu, L.N, Liu, L, Wang, F. | | Deposit date: | 2019-06-17 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Met1-specific motifs conserved in OTUB subfamily of green plants enable rice OTUB1 to hydrolyse Met1 ubiquitin chains
Nat Commun, 13, 2022
|
|
6KBE
 
 | | Structure of Deubiquitinase | | Descriptor: | Polyubiquitin-C, Ubiquitin thioesterase | | Authors: | Lu, L.N, Liu, L, Wang, F. | | Deposit date: | 2019-06-24 | | Release date: | 2020-06-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.339 Å) | | Cite: | Met1-specific motifs conserved in OTUB subfamily of green plants enable rice OTUB1 to hydrolyse Met1 ubiquitin chains
Nat Commun, 13, 2022
|
|
6K9P
 
 | | Structure of Deubiquitinase | | Descriptor: | Ubiquitin, Ubiquitin thioesterase | | Authors: | Lu, L.N, Liu, L, Wang, F. | | Deposit date: | 2019-06-17 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.047 Å) | | Cite: | Met1-specific motifs conserved in OTUB subfamily of green plants enable rice OTUB1 to hydrolyse Met1 ubiquitin chains
Nat Commun, 13, 2022
|
|
6OVB
 
 | | Crystal structure of a Bacillus thuringiensis Cry1Da tryptic core variant | | Descriptor: | Active core crystal toxin protein 1D | | Authors: | Rydel, T.J, Halls, C, Evdokimov, A.G, Beishir, S.C. | | Deposit date: | 2019-05-07 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.611 Å) | | Cite: | Bacillus thuringiensis Cry1Da_7 and Cry1B.868 Protein Interactions with Novel Receptors Allow Control of Resistant Fall Armyworms, Spodoptera frugiperda (J.E. Smith).
Appl.Environ.Microbiol., 85, 2019
|
|
6OWK
 
 | |
6J1O
 
 | |
6J46
 
 | | LepI-SAH complex structure | | Descriptor: | O-methyltransferase lepI, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Qiu, S, Wei, C. | | Deposit date: | 2019-01-08 | | Release date: | 2019-05-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.621 Å) | | Cite: | Deciphering the regulatory and catalytic mechanisms of an unusual SAM-dependent enzyme.
Signal Transduct Target Ther, 4, 2019
|
|
6J24
 
 | | Crystal structure of a SAM-dependent methyltransferase LepI in complex with its substrate | | Descriptor: | (3~{S},4'~{R},4'~{a}~{S},6'~{R},8'~{a}~{S})-4',6'-dimethyl-5-phenyl-spiro[1~{H}-pyridine-3,5'-2,3,4,4~{a},6,8~{a}-hexahydro-1~{H}-naphthalene]-2,4-dione, O-methyltransferase, S-ADENOSYLMETHIONINE | | Authors: | Qiu, S, Wei, C. | | Deposit date: | 2018-12-30 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.244 Å) | | Cite: | Deciphering the regulatory and catalytic mechanisms of an unusual SAM-dependent enzyme.
Signal Transduct Target Ther, 4, 2019
|
|
7RN2
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with SJ001010551-2 | | Descriptor: | 1,2-ETHANEDIOL, 2-[(6S,10S)-4-(4-chlorophenyl)-2,3,9-trimethyl-6H-thieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-6-yl]-N-[(pyridin-2-yl)methyl]acetamide, Bromodomain-containing protein 4 | | Authors: | Stachowski, T.R, Fischer, M. | | Deposit date: | 2021-07-28 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | From PROTAC to inhibitor: Structure-guided discovery of potent and orally bioavailable BET inhibitors.
Eur.J.Med.Chem., 251, 2023
|
|
7RMD
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with SJ001011461-1 | | Descriptor: | 1,2-ETHANEDIOL, 2-[(6S,10R)-4-(4-chlorophenyl)-2,3,9-trimethyl-6H-thieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-6-yl]-N-(1,3,4-thiadiazol-2-yl)acetamide, Bromodomain-containing protein 4 | | Authors: | Stachowski, T.R, Fischer, M. | | Deposit date: | 2021-07-27 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | From PROTAC to inhibitor: Structure-guided discovery of potent and orally bioavailable BET inhibitors.
Eur.J.Med.Chem., 251, 2023
|
|
