2C4U
 
 | | Crystal structure of the apo form of the 5'-Fluoro-5'-deoxyadenosine synthase enzyme from Streptomyces cattleya | | Descriptor: | 5'-FLUORO-5'-DEOXYADENOSINE SYNTHASE, GLYCEROL | | Authors: | McEwan, A.R, Deng, H, Robinson, D.A, DeLaurentis, W, McGlinchey, R.P, O'Hagan, D, Naismith, J.H. | | Deposit date: | 2005-10-22 | | Release date: | 2006-04-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Substrate specificity in enzymatic fluorination. The fluorinase from Streptomyces cattleya accepts 2'-deoxyadenosine substrates.
Org. Biomol. Chem., 4, 2006
|
|
3VCB
 
 | | C425S mutant of the C-terminal cytoplasmic domain of non-structural protein 4 from mouse hepatitis virus A59 | | Descriptor: | RNA-directed RNA polymerase | | Authors: | Xu, X, Lou, Z, Ma, Y, Chen, X, Yang, Z, Tong, X, Zhao, Q, Xu, Y, Deng, H, Bartlam, M, Rao, Z. | | Deposit date: | 2012-01-03 | | Release date: | 2012-01-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the C-terminal cytoplasmic domain of non-structural protein 4 from mouse hepatitis virus A59.
Plos One, 4, 2009
|
|
3VC8
 
 | | Crystal structure of the C-terminal cytoplasmic domain of non-structural protein 4 from mouse hepatitis virus A59 | | Descriptor: | RNA-directed RNA polymerase | | Authors: | Xu, X, Lou, Z, Ma, Y, Chen, X, Yang, Z, Tong, X, Zhao, Q, Xu, Y, Deng, H, Bartlam, M, Rao, Z. | | Deposit date: | 2012-01-03 | | Release date: | 2012-01-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the C-terminal cytoplasmic domain of non-structural protein 4 from mouse hepatitis virus A59.
Plos One, 4, 2009
|
|
5LMZ
 
 | | Fluorinase from Streptomyces sp. MA37 | | Descriptor: | 1-DEAZA-ADENOSINE, CHLORIDE ION, Fluorinase, ... | | Authors: | Mann, G, O'Hagan, D, Naismith, J.H, Bandaranayaka, N. | | Deposit date: | 2016-08-02 | | Release date: | 2016-08-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Identification of fluorinases from Streptomyces sp MA37, Norcardia brasiliensis, and Actinoplanes sp N902-109 by genome mining.
Chembiochem, 15, 2014
|
|
2C2W
 
 | |
2FDA
 
 | | Crystal Structure of the Catalytic Domain of Human Coagulation Factor XIa in Complex with alpha-Ketothiazole Arginine Derived Ligand | | Descriptor: | BICARBONATE ION, Coagulation factor XI, N~2~-(AMINOCARBONYL)-N~1~-{4-{[AMINO(IMINO)METHYL]AMINO}-1-[HYDROXY(1,3-THIAZOL-2-YL)METHYL]BUTYL}VALINAMIDE, ... | | Authors: | Jin, L, Pandey, P, Babine, R.E, Weaver, D.T, Abdel-Meguid, S.S, Strickler, J.E. | | Deposit date: | 2005-12-13 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Synthesis, SAR exploration, and X-ray crystal structures of factor XIa inhibitors containing an alpha-ketothiazole arginine
Bioorg.Med.Chem.Lett., 16, 2006
|
|
5HNE
 
 | | X-RAY CRYSTAL STRUCTURE OF HUMAN MITOCHONDRIAL BRANCHED CHAIN AMINOTRANSFERASE (BCATM) COMPLEXED WITH A 2-ARYL BENZIMIDAZOLE COMPOUND AND AN INTERNAL ALDIMINE LINKED PLP COFACTOR | | Descriptor: | 1,2-ETHANEDIOL, 1-[(1R,3S)-3-{[(5-bromothiophen-2-yl)carbonyl]amino}cyclohexyl]-N-methyl-2-(pyridin-2-yl)-1H-benzimidazole-5-carboxamide, Branched-chain-amino-acid aminotransferase, ... | | Authors: | Somers, D.O. | | Deposit date: | 2016-01-18 | | Release date: | 2016-05-04 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Discovery and Optimization of Potent, Selective, and in Vivo Efficacious 2-Aryl Benzimidazole BCATm Inhibitors.
Acs Med.Chem.Lett., 7, 2016
|
|
5CR5
 
 | |
8XFB
 
 | | Cryo-EM structure of partial dimeric WDR11-FAM91A1 complex | | Descriptor: | Protein FAM91A1, WD repeat-containing protein 11 | | Authors: | Jia, G.W, Deng, Q.H, Su, Z.M, Jia, D. | | Deposit date: | 2023-12-13 | | Release date: | 2024-08-14 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The WDR11 complex is a receptor for acidic-cluster-containing cargo proteins.
Cell, 187, 2024
|
|
8Z9M
 
 | | Cryo-EM structure of dimeric WDR11-FAM91A1 complex | | Descriptor: | Protein FAM91A1, WD repeat-containing protein 11 | | Authors: | Jia, G.W, Deng, Q.H, Su, Z.M, Jia, D. | | Deposit date: | 2024-04-23 | | Release date: | 2024-08-14 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The WDR11 complex is a receptor for acidic-cluster-containing cargo proteins.
Cell, 187, 2024
|
|
7BYF
 
 | | The crystal structure of mouse ORF10-Rae1-Nup98 complex | | Descriptor: | 10 protein, MERCURY (II) ION, Peptidase S59 domain-containing protein, ... | | Authors: | Gao, P, Feng, H. | | Deposit date: | 2020-04-22 | | Release date: | 2021-03-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular mechanism underlying selective inhibition of mRNA nuclear export by herpesvirus protein ORF10.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3LT3
 
 | | Crystal structure of Rv3671c from M. tuberculosis H37Rv, Ser343Ala mutant, inactive form | | Descriptor: | POSSIBLE MEMBRANE-ASSOCIATED SERINE PROTEASE | | Authors: | Biswas, T, Small, J, Vandal, O, Ehrt, S, Tsodikov, O.V. | | Deposit date: | 2010-02-14 | | Release date: | 2010-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insight into serine protease Rv3671c that Protects M. tuberculosis from oxidative and acidic stress.
Structure, 18, 2010
|
|
4Y5O
 
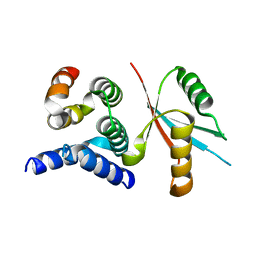 | | CCM2 HHD in complex with MEKK3 NPB1 | | Descriptor: | Malcavernin, Mitogen-activated protein kinase kinase kinase 3 | | Authors: | Fisher, O.S, Boggon, T.J. | | Deposit date: | 2015-02-11 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure and vascular function of MEKK3-cerebral cavernous malformations 2 complex.
Nat Commun, 6, 2015
|
|
4DNW
 
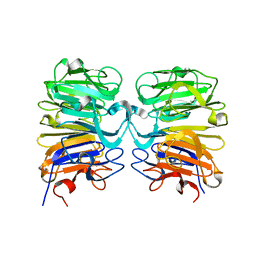 | | Crystal structure of UVB-resistance protein UVR8 | | Descriptor: | AT5g63860/MGI19_6 | | Authors: | Wu, D, Hu, Q, Yan, Z, Chen, W, Yan, C, Wang, J, Shi, Y. | | Deposit date: | 2012-02-09 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.773 Å) | | Cite: | Structural basis of ultraviolet-B perception by UVR8.
Nature, 484, 2012
|
|
4DNV
 
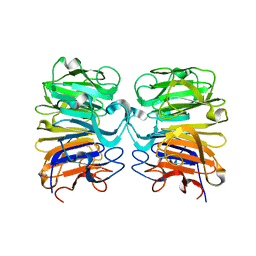 | | Crystal structure of the W285F mutant of UVB-resistance protein UVR8 | | Descriptor: | AT5g63860/MGI19_6 | | Authors: | Wu, D, Hu, Q, Yan, Z, Chen, W, Yan, C, Zhang, J, Wang, J, Shi, Y. | | Deposit date: | 2012-02-09 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Structural basis of ultraviolet-B perception by UVR8.
Nature, 484, 2012
|
|
4DNU
 
 | | Crystal structure of the W285A mutant of UVB-resistance protein UVR8 | | Descriptor: | AT5g63860/MGI19_6 | | Authors: | Wu, D, Hu, Q, Yan, Z, Chen, W, Yan, C, Zhang, J, Wang, J, Shi, Y. | | Deposit date: | 2012-02-09 | | Release date: | 2012-03-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.764 Å) | | Cite: | Structural basis of ultraviolet-B perception by UVR8.
Nature, 484, 2012
|
|
4P1U
 
 | |
6K15
 
 | | RSC substrate-recruitment module | | Descriptor: | Chromatin structure-remodeling complex protein RSC3, Chromatin structure-remodeling complex protein RSC30, Chromatin structure-remodeling complex protein RSC58, ... | | Authors: | Ye, Y.P, Wu, H, Chen, K.J, Verma, N, Cairns, B, Gao, N, Chen, Z.C. | | Deposit date: | 2019-05-09 | | Release date: | 2019-11-13 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the RSC complex bound to the nucleosome.
Science, 366, 2019
|
|
8AQF
 
 | | CRYSTAL STRUCTURE OF HUMAN MONOGLYCERIDE LIPASE WITH COMPOUND LEI-515 | | Descriptor: | 1-[(~{R})-[2-chloranyl-4-[(2~{S},3~{S})-4-(3-chlorophenyl)-2,3-dimethyl-piperazin-1-yl]carbonyl-phenyl]sulfinyl]-3,3-bis(fluoranyl)pentan-2-one, Monoglyceride lipase | | Authors: | Jiang, M, Huizenga, M, Wirt, J, Paloczi, J, Amedi, A, van der Berg, R, Benz, J, Collin, L, Deng, H, Driever, W, Florea, B, Grether, U, Janssen, A, Heitman, L, Lam, T.W, Mohr, F, Pavlovic, A, Ruf, I, Rutjes, H, Stevens, F, van der Vliet, D, van der Wel, T, Wittwer, M, Boeckel, C, Pacher, P, Hohmann, A, van der Stelt, M. | | Deposit date: | 2022-08-12 | | Release date: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of a peripheral restricted, reversible monoacylglycerol lipase inhibitor that reduces liver injury and chemotherapy-induced neuropathy
To Be Published
|
|
3H6F
 
 | |
8JJ9
 
 | |
2OYP
 
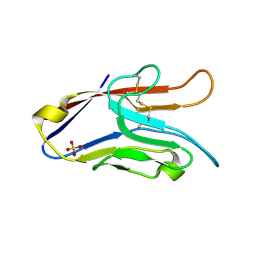 | | T Cell Immunoglobulin Mucin-3 Crystal Structure Revealed a Galectin-9-independent Binding Surface | | Descriptor: | Hepatitis A virus cellular receptor 2, SULFATE ION | | Authors: | Cao, E, Ramagopal, U.A, Fedorov, A.A, Fedorov, E.V, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2007-02-22 | | Release date: | 2007-04-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | T cell immunoglobulin mucin-3 crystal structure reveals a galectin-9-independent ligand-binding surface
Immunity, 26, 2007
|
|
3HFA
 
 | |
3HF9
 
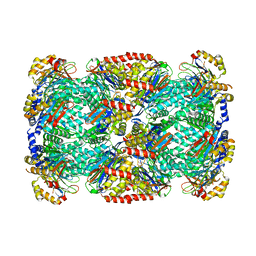 | |
2OA5
 
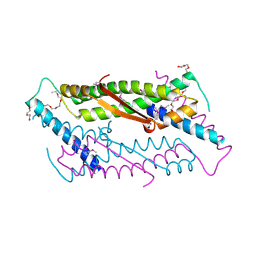 | | Crystal structure of ORF52 from Murid herpesvirus (MUHV-4) (Murine gammaherpesvirus 68) at 2.1 A resolution. Northeast Structural Genomics Consortium target MHR28B. | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, Hypothetical protein BQLF2 | | Authors: | Benach, J, Chen, Y, Seetharaman, J, Janjua, H, Xiao, R, Cunningham, K, Ma, L.-C, Ho, C.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-12-14 | | Release date: | 2007-01-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and functional studies of the abundant tegument protein ORF52 from murine gammaherpesvirus 68.
J.Biol.Chem., 282, 2007
|
|
