6JZ7
 
 | |
6JZ6
 
 | | b-glucuronidase from Ruminococcus gnavus in complex with C6-substituted uronic isofagomine | | Descriptor: | (2~{S},3~{S},4~{R},5~{R})-4,5-bis(oxidanyl)-2-propyl-piperidine-3-carboxylic acid, Beta-glucuronidase | | Authors: | Dashnyam, P, Lin, H.Y. | | Deposit date: | 2019-04-30 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.605 Å) | | Cite: | Substituent Position of Iminocyclitols Determines the Potency and Selectivity for Gut Microbial Xenobiotic-Reactivating Enzymes.
J.Med.Chem., 63, 2020
|
|
6JZ8
 
 | | b-glucuronidase from Ruminococcus gnavus in complex with D-glucaro 1,5-lactone | | Descriptor: | (2S,3S,4S,5R)-3,4,5-trihydroxy-6-oxo-oxane-2-carboxylic acid, Beta-glucuronidase | | Authors: | Dashnyam, P, Lin, H.Y. | | Deposit date: | 2019-04-30 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.583 Å) | | Cite: | Substituent Position of Iminocyclitols Determines the Potency and Selectivity for Gut Microbial Xenobiotic-Reactivating Enzymes.
J.Med.Chem., 63, 2020
|
|
6JZ5
 
 | |
6JZ3
 
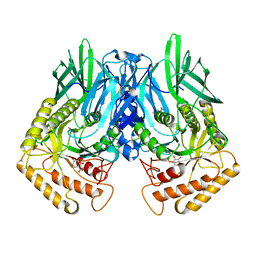 | | b-glucuronidase from Ruminococcus gnavus in complex with uronic deoxynojirimycin | | Descriptor: | (2~{S},3~{R},4~{R},5~{S})-3,4,5-tris(oxidanyl)piperidine-2-carboxylic acid, (4R)-2-METHYLPENTANE-2,4-DIOL, Beta-glucuronidase | | Authors: | Dashnyam, P, Lin, H.Y. | | Deposit date: | 2019-04-30 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Substituent Position of Iminocyclitols Determines the Potency and Selectivity for Gut Microbial Xenobiotic-Reactivating Enzymes.
J.Med.Chem., 63, 2020
|
|
7D5F
 
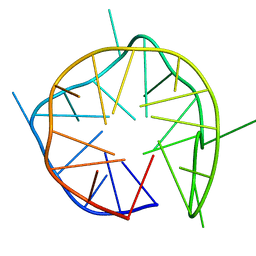 | | Left-handed G-quadruplex containing 3 bulges | | Descriptor: | 3xBulge-LHG4motif | | Authors: | Winnerdy, F.R, Das, P, Ngo, K.H, Maity, A, Bakalar, B, Mechulam, Y, Schmitt, E, Phan, A.T. | | Deposit date: | 2020-09-25 | | Release date: | 2021-02-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Bulges in left-handed G-quadruplexes.
Nucleic Acids Res., 49, 2021
|
|
4IGH
 
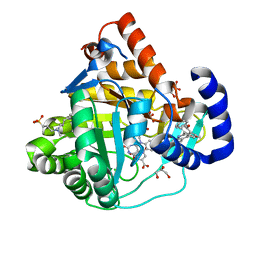 | | High resolution crystal structure of human dihydroorotate dehydrogenase bound with 4-quinoline carboxylic acid analog | | Descriptor: | 3-[decyl(dimethyl)ammonio]propane-1-sulfonate, 6-fluoro-2-[2-methyl-4-phenoxy-5-(propan-2-yl)phenyl]quinoline-4-carboxylic acid, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Deng, X, Das, P, Fontoura, B.M.A, Phillips, M.A, De Brabander, J.K. | | Deposit date: | 2012-12-17 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | SAR Based Optimization of a 4-Quinoline Carboxylic Acid Analog with Potent Anti-Viral Activity.
ACS MED.CHEM.LETT., 4, 2013
|
|
8Z2F
 
 | |
8Z1M
 
 | |
8Z1N
 
 | |
8Z1C
 
 | |
8Z1O
 
 | |
6SC4
 
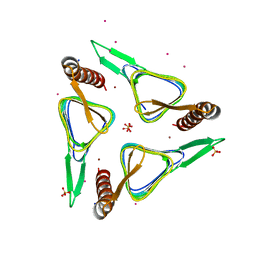 | | Gamma-Carbonic Anhydrase from the Haloarchaeon Halobacterium sp. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, CADMIUM ION, ... | | Authors: | Vogler, M, Karan, R, Renn, D, Vancea, A, Vielberg, V.-T, Groetzinger, S.W, DasSarma, P, Das Sarma, S, Eppinger, J, Groll, M, Rueping, M. | | Deposit date: | 2019-07-23 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure and Active Site Engineering of a Halophilic gamma-Carbonic Anhydrase.
Front Microbiol, 11, 2020
|
|
1A0N
 
 | | NMR STUDY OF THE SH3 DOMAIN FROM FYN PROTO-ONCOGENE TYROSINE KINASE COMPLEXED WITH THE SYNTHETIC PEPTIDE P2L CORRESPONDING TO RESIDUES 91-104 OF THE P85 SUBUNIT OF PI3-KINASE, FAMILY OF 25 STRUCTURES | | Descriptor: | FYN, PRO-PRO-ARG-PRO-LEU-PRO-VAL-ALA-PRO-GLY-SER-SER-LYS-THR | | Authors: | Renzoni, D.A, Pugh, D.J.R, Siligardi, G, Das, P, Morton, C.J, Rossi, C, Waterfield, M.D, Campbell, I.D, Ladbury, J.E. | | Deposit date: | 1997-12-05 | | Release date: | 1998-02-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural and thermodynamic characterization of the interaction of the SH3 domain from Fyn with the proline-rich binding site on the p85 subunit of PI3-kinase.
Biochemistry, 35, 1996
|
|
1AZG
 
 | | NMR STUDY OF THE SH3 DOMAIN FROM FYN PROTO-ONCOGENE TYROSINE KINASE KINASE COMPLEXED WITH THE SYNTHETIC PEPTIDE P2L CORRESPONDING TO RESIDUES 91-104 OF THE P85 SUBUNIT OF PI3-KINASE, MINIMIZED AVERAGE (PROBMAP) STRUCTURE | | Descriptor: | FYN, PRO-PRO-ARG-PRO-LEU-PRO-VAL-ALA-PRO-GLY-SER-SER-LYS-THR | | Authors: | Renzoni, D.A, Pugh, D.J.R, Siligardi, G, Das, P, Morton, C.J, Rossi, C, Waterfield, M.D, Campbell, I.D, Ladbury, J.E. | | Deposit date: | 1997-11-18 | | Release date: | 1998-02-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural and thermodynamic characterization of the interaction of the SH3 domain from Fyn with the proline-rich binding site on the p85 subunit of PI3-kinase.
Biochemistry, 35, 1996
|
|
1D9E
 
 | | STRUCTURE OF E. COLI KDO8P SYNTHASE | | Descriptor: | 3-DEOXY-D-MANNO-OCTULOSONATE 8-PHOSPHATE SYNTHASE, SULFATE ION | | Authors: | Radaev, S, Dastidar, P, Patel, M, Woodard, R.W, Gatti, D.L. | | Deposit date: | 1999-10-27 | | Release date: | 2000-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and mechanism of 3-deoxy-D-manno-octulosonate 8-phosphate synthase.
J.Biol.Chem., 275, 2000
|
|
5ZH4
 
 | | CRYSTAL STRUCTURE OF PfKRS WITH INHIBITOR CLADO-7 | | Descriptor: | (3R)-6,8-dihydroxy-3-{[(2S,6R)-6-methyloxan-2-yl]methyl}-3,4-dihydro-1H-2-benzopyran-1-one, CHLORIDE ION, LYSINE, ... | | Authors: | Babbar, P, Malhotra, N, Sharma, M, Harlos, K, Reddy, D.S, Manickam, Y, Sharma, A. | | Deposit date: | 2018-03-11 | | Release date: | 2018-06-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Specific Stereoisomeric Conformations Determine the Drug Potency of Cladosporin Scaffold against Malarial Parasite
J. Med. Chem., 61, 2018
|
|
5ZH3
 
 | | CRYSTAL STRUCTURE OF PfKRS WITH INHIBITOR CLADO-6 | | Descriptor: | (3S)-6,8-dihydroxy-3-{[(2R,6R)-6-methyloxan-2-yl]methyl}-3,4-dihydro-1H-2-benzopyran-1-one, LYSINE, Lysine-tRNA ligase | | Authors: | Babbar, P, Malhotra, N, Sharma, M, Harlos, K, Reddy, D.S, Manickam, Y, Sharma, A. | | Deposit date: | 2018-03-11 | | Release date: | 2018-06-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Specific Stereoisomeric Conformations Determine the Drug Potency of Cladosporin Scaffold against Malarial Parasite
J. Med. Chem., 61, 2018
|
|
5ZH2
 
 | | CRYSTAL STRUCTURE OF PfKRS WITH INHIBITOR CLADO-5 | | Descriptor: | (3R)-6,8-dihydroxy-3-{[(2R,6R)-6-methyloxan-2-yl]methyl}-3,4-dihydro-1H-2-benzopyran-1-one, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, ... | | Authors: | Babbar, P, Malhotra, N, Sharma, M, Harlos, K, Reddy, D.S, Manickam, Y, Sharma, A. | | Deposit date: | 2018-03-11 | | Release date: | 2018-06-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Specific Stereoisomeric Conformations Determine the Drug Potency of Cladosporin Scaffold against Malarial Parasite
J. Med. Chem., 61, 2018
|
|
5ZH5
 
 | | CRYSTAL STRUCTURE OF PfKRS WITH INHIBITOR CLADO-2 | | Descriptor: | (3S)-6,8-dihydroxy-3-{[(2R,6S)-6-methyloxan-2-yl]methyl}-3,4-dihydro-1H-2-benzopyran-1-one, CHLORIDE ION, LYSINE, ... | | Authors: | Babbar, P, Malhotra, N, Sharma, M, Harlos, K, Reddy, D.S, Manickam, Y, Sharma, A. | | Deposit date: | 2018-03-11 | | Release date: | 2018-06-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Specific Stereoisomeric Conformations Determine the Drug Potency of Cladosporin Scaffold against Malarial Parasite
J. Med. Chem., 61, 2018
|
|
8Z2A
 
 | |
8Z2B
 
 | |
8Z29
 
 | |
8Z2C
 
 | |
8XAK
 
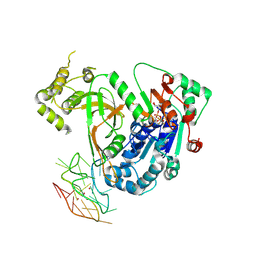 | | Structure of Pif1-G4 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase PIF1, DNA (34-MER), ... | | Authors: | Hong, Z, Song, H. | | Deposit date: | 2023-12-04 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Eukaryotic Pif1 helicase unwinds G-quadruplex and dsDNA using a conserved wedge.
Nat Commun, 15, 2024
|
|
