2YBS
 
 | |
2YDE
 
 | | FACTOR INHIBITING HIF-1 ALPHA IN COMPLEX WITH S-2-HYDROXYGLUTARATE | | Descriptor: | (2S)-2-HYDROXYPENTANEDIOIC ACID, FE (III) ION, GLYCEROL, ... | | Authors: | Chowdhury, R, Clifton, I.J, Schofield, C.J. | | Deposit date: | 2011-03-18 | | Release date: | 2011-03-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases.
EMBO Rep., 12, 2011
|
|
2YBP
 
 | | JMJD2A COMPLEXED WITH R-2-HYDROXYGLUTARATE AND HISTONE H3K36me3 PEPTIDE (30-41) | | Descriptor: | (2R)-2-hydroxypentanedioic acid, GLYCEROL, HISTONE H3.1T, ... | | Authors: | Chowdhury, R, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2011-03-09 | | Release date: | 2011-03-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases.
EMBO Rep., 12, 2011
|
|
2YBK
 
 | | JMJD2A COMPLEXED WITH R-2-HYDROXYGLUTARATE | | Descriptor: | (2R)-2-hydroxypentanedioic acid, CHLORIDE ION, LYSINE-SPECIFIC DEMETHYLASE 4A, ... | | Authors: | Chowdhury, R, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2011-03-08 | | Release date: | 2011-04-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases.
EMBO Rep., 12, 2011
|
|
4PVO
 
 | | Crystal Structure of VIM-2 metallo-beta-lactamase in complex with ML302 and ML302F | | Descriptor: | (2Z)-2-sulfanyl-3-(2,3,6-trichlorophenyl)prop-2-enoic acid, Beta-lactamase class B VIM-2, DIMETHYL SULFOXIDE, ... | | Authors: | Aik, W.S, Brem, J, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2014-03-18 | | Release date: | 2014-11-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Rhodanine hydrolysis leads to potent thioenolate mediated metallo-beta-lactamase inhibition.
Nat.Chem., 6, 2014
|
|
4PVT
 
 | | Crystal Structure of VIM-2 metallo-beta-lactamase in complex with ML302F | | Descriptor: | (2Z)-2-sulfanyl-3-(2,3,6-trichlorophenyl)prop-2-enoic acid, Beta-lactamase class B VIM-2, FORMIC ACID, ... | | Authors: | Aik, W.S, Brem, J, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2014-03-18 | | Release date: | 2014-11-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Rhodanine hydrolysis leads to potent thioenolate mediated metallo-beta-lactamase inhibition.
Nat.Chem., 6, 2014
|
|
5OX5
 
 | | HIF prolyl hydroxylase 2 (PHD2/ EGLN1) in complex with CCT6, a GSK1278863-related compound | | Descriptor: | (6-hydroxy-1,3-dimethyl-2,4-dioxo-1,2,3,4-tetrahydropyrimidine-5-carbonyl)glycine, BICARBONATE ION, Egl nine homolog 1, ... | | Authors: | Chowdhury, R, Thinnes, C.C, Schofield, C.J. | | Deposit date: | 2017-09-06 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Molecular and cellular mechanisms of HIF prolyl hydroxylase inhibitors in clinical trials.
Chem Sci, 8, 2017
|
|
5OX6
 
 | | HIF prolyl hydroxylase 2 (PHD2/ EGLN1) in complex with Vadadustat | | Descriptor: | Egl nine homolog 1, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Chowdhury, R, Zhang, D, Schofield, C.J. | | Deposit date: | 2017-09-06 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Molecular and cellular mechanisms of HIF prolyl hydroxylase inhibitors in clinical trials.
Chem Sci, 8, 2017
|
|
4KQQ
 
 | | CRYSTAL STRUCTURE OF PENICILLIN-BINDING PROTEIN 3 FROM PSEUDOMONAS AERUGINOSA IN COMPLEX WITH (5S)-Penicilloic Acid | | Descriptor: | (2S,4S)-2-[(R)-carboxy{[(2R)-2-{[(4-ethyl-2,3-dioxopiperazin-1-yl)carbonyl]amino}-2-phenylacetyl]amino}methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Nettleship, J.E, Stuart, D.I, Owens, R.J, Ren, J. | | Deposit date: | 2013-05-15 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Binding of (5S)-Penicilloic Acid to Penicillin Binding Protein 3.
Acs Chem.Biol., 8, 2013
|
|
4KQR
 
 | | CRYSTAL STRUCTURE OF PENICILLIN-BINDING PROTEIN 3 FROM PSEUDOMONAS AERUGINOSA IN COMPLEX WITH (5S)-Penicilloic Acid | | Descriptor: | (2S,4S)-2-[(R)-carboxy{[(2R)-2-{[(4-ethyl-2,3-dioxopiperazin-1-yl)carbonyl]amino}-2-phenylacetyl]amino}methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Nettleship, J.E, Stuart, D.I, Owens, R.J, Ren, J. | | Deposit date: | 2013-05-15 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Binding of (5S)-Penicilloic Acid to Penicillin Binding Protein 3.
Acs Chem.Biol., 8, 2013
|
|
4KQO
 
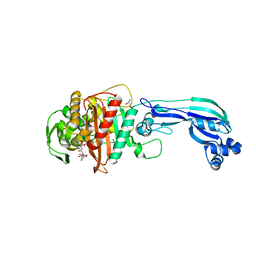 | | Crystal structure of penicillin-binding protein 3 from pseudomonas aeruginosa in complex with piperacillin | | Descriptor: | CHLORIDE ION, GLYCEROL, IMIDAZOLE, ... | | Authors: | Nettleship, J.E, Stuart, D.I, Owens, R.J, Ren, J. | | Deposit date: | 2013-05-15 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Binding of (5S)-Penicilloic Acid to Penicillin Binding Protein 3.
Acs Chem.Biol., 8, 2013
|
|
5LS3
 
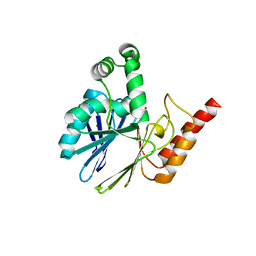 | |
6I7R
 
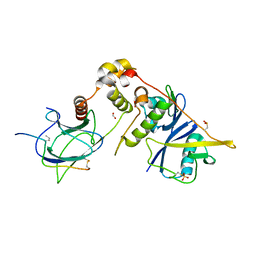 | |
6I7Q
 
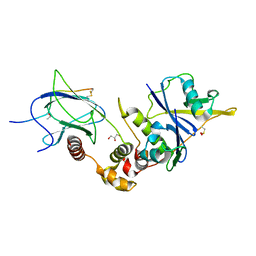 | |
4B7K
 
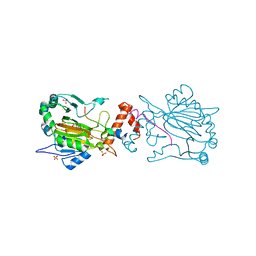 | | FACTOR INHIBITING HIF-1 ALPHA IN COMPLEX WITH CONSENSUS ANKYRIN REPEAT DOMAIN-SER PEPTIDE (20-MER) | | Descriptor: | CONSENSUS ANKYRIN REPEAT DOMAIN-SER, HYPOXIA-INDUCIBLE FACTOR 1-ALPHA INHIBITOR, N-OXALYLGLYCINE, ... | | Authors: | Chowdhury, R, Ge, W, Schofield, C.J. | | Deposit date: | 2012-08-20 | | Release date: | 2013-01-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Substrate selectivity analyses of factor inhibiting hypoxia-inducible factor.
Angew. Chem. Int. Ed. Engl., 52, 2013
|
|
4B7E
 
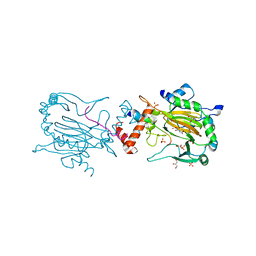 | | FACTOR INHIBITING HIF-1 ALPHA IN COMPLEX WITH CONSENSUS ANKYRIN REPEAT DOMAIN-LEU PEPTIDE (20-MER) | | Descriptor: | CONSENSUS ANKYRIN REPEAT DOMAIN-LEU, GLYCEROL, HYPOXIA-INDUCIBLE FACTOR 1-ALPHA INHIBITOR, ... | | Authors: | Chowdhury, R, Ge, W, Schofield, C.J. | | Deposit date: | 2012-08-17 | | Release date: | 2013-01-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Substrate selectivity analyses of factor inhibiting hypoxia-inducible factor.
Angew. Chem. Int. Ed. Engl., 52, 2013
|
|
4CWD
 
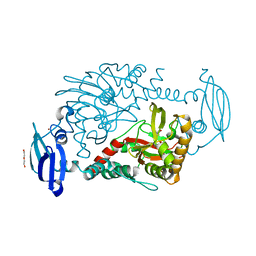 | | CRYSTAL STRUCTURE OF HUMAN GAMMA-BUTYROBETAINE,2-OXOGLUTARATE IN COMPLEX WITH 449, A NOVEL SUBSTRATE | | Descriptor: | 4-carboxy-1,1-dimethylpiperidin-1-ium, DI(HYDROXYETHYL)ETHER, GAMMA-BUTYROBETAINE DIOXYGENASE, ... | | Authors: | McDonough, M.A, Kochan, G, Rydzik, A, Schofield, C.J. | | Deposit date: | 2014-04-02 | | Release date: | 2014-09-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Oxygenase-catalyzed desymmetrization of N,N-dialkyl-piperidine-4-carboxylic acids.
Angew. Chem. Int. Ed. Engl., 53, 2014
|
|
4BP0
 
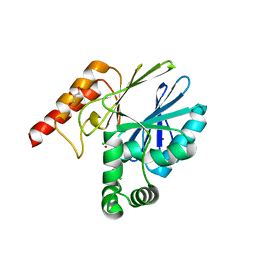 | | Crystal structure of the closed form of Pseudomonas aeruginosa SPM-1 | | Descriptor: | CHLORIDE ION, GLYCEROL, METALLO-B-LACTAMASE, ... | | Authors: | McDonough, M.A, Brem, J, Schofield, C.J. | | Deposit date: | 2013-05-22 | | Release date: | 2014-06-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Studying the active-site loop movement of the Sao Paolo metallo-beta-lactamase-1
Chem Sci, 6, 2015
|
|
5L9V
 
 | |
5LAS
 
 | |
5J5Y
 
 | | Translation initiation factor 4E in complex with m2(7,2'O)GppCCl2ppG mRNA 5' cap analog | | Descriptor: | 2-amino-9-{5-O-[(R)-{[(S)-{dichloro[(R)-hydroxy(phosphonooxy)phosphoryl]methyl}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]-2-O-methyl-beta-D-ribofuranosyl}-7-methyl-9H-purin-7-ium-6-olate, Eukaryotic translation initiation factor 4E, GLYCEROL | | Authors: | Warminski, M, Nowak, E, Rydzik, A.M, Kowalska, J, Jemielity, J, Nowotny, M. | | Deposit date: | 2016-04-04 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | mRNA cap analogues substituted in the tetraphosphate chain with CX2: identification of O-to-CCl2 as the first bridging modification that confers resistance to decapping without impairing translation.
Nucleic Acids Res., 45, 2017
|
|
5J5O
 
 | | Translation initiation factor 4E in complex with m7GppppG mRNA 5' cap analog | | Descriptor: | 5'-O-[(R)-hydroxy{[(R)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]oxy}phosphoryl]-7-methylguanosine, Eukaryotic translation initiation factor 4E, GLYCEROL | | Authors: | Warminski, M, Nowak, E, Rydzik, A.M, Kowalska, J, Jemielity, J, Nowotny, M. | | Deposit date: | 2016-04-03 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.867 Å) | | Cite: | mRNA cap analogues substituted in the tetraphosphate chain with CX2: identification of O-to-CCl2 as the first bridging modification that confers resistance to decapping without impairing translation.
Nucleic Acids Res., 45, 2017
|
|
5LB6
 
 | |
5LBE
 
 | |
5LBC
 
 | |
