7UEN
 
 | |
7UEM
 
 | |
4X4Z
 
 | |
6ARP
 
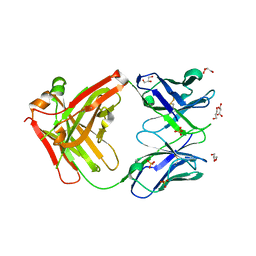 | | Structure of a mutant Cetuximab Fab fragment | | Descriptor: | Cetuximab mutant Fab heavy chain,Immunoglobulin gamma-1 heavy chain, Cetuximab mutant light chain,Uncharacterized protein, GLYCEROL, ... | | Authors: | Christie, M, Christ, D. | | Deposit date: | 2017-08-23 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a mutant Cetuximab Fab fragment
To Be Published
|
|
4N1E
 
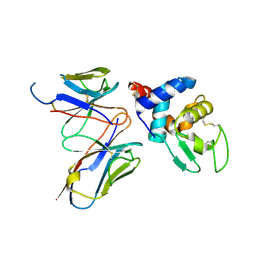 | | Structural evidence for antigen receptor evolution | | Descriptor: | Lysozyme C, immunoglobulin variable light chain domain | | Authors: | Langley, D.B, Rouet, R, Stock, D, Christ, D. | | Deposit date: | 2013-10-04 | | Release date: | 2014-10-29 | | Last modified: | 2018-04-18 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structural reconstruction of protein ancestry.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4N1C
 
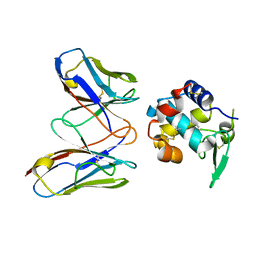 | | Structural evidence for antigen receptor evolution | | Descriptor: | Lysozyme C, immunoglobulin variable light chain domain | | Authors: | Langley, D.B, Rouet, R, Roome, B, Stock, D, Christ, D. | | Deposit date: | 2013-10-03 | | Release date: | 2014-10-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural reconstruction of protein ancestry.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8DXT
 
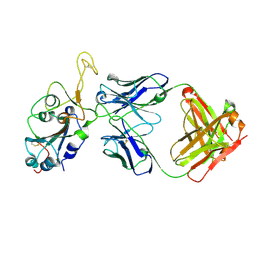 | | Fab arm of antibody GAR12 bound to the receptor binding domain of SARS-CoV-2. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab arm of antibody GAR12, Light chain of Fab arm of antibody GAR12, ... | | Authors: | Langley, D.B, Christ, D, Henry, J.Y. | | Deposit date: | 2022-08-03 | | Release date: | 2023-01-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Broadly neutralizing SARS-CoV-2 antibodies through epitope-based selection from convalescent patients.
Nat Commun, 14, 2023
|
|
5VJQ
 
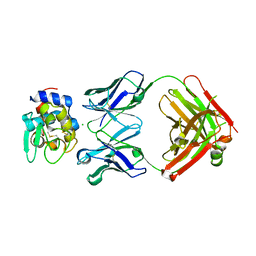 | | Complex between HyHEL10 Fab fragment heavy chain mutant (I29F, S52T, Y53F) and Pekin duck egg lysozyme isoform I (DEL-I) | | Descriptor: | CHLORIDE ION, GLYCEROL, HyHEL10 heavy chain Fab fragment carrying three mutations; I29F, ... | | Authors: | Langley, D.B, Christ, D. | | Deposit date: | 2017-04-19 | | Release date: | 2018-04-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Germinal center antibody mutation trajectories are determined by rapid self/foreign discrimination.
Science, 360, 2018
|
|
7T72
 
 | | Epitope-based selection of SARS-CoV-2 neutralizing antibodies from convalescent patients | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody heavy chain, Antibody light chain, ... | | Authors: | Langley, D.B, Christ, D, Rouet, R. | | Deposit date: | 2021-12-14 | | Release date: | 2022-12-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.177 Å) | | Cite: | Broadly neutralizing SARS-CoV-2 antibodies through epitope-based selection from convalescent patients.
Nat Commun, 14, 2023
|
|
8CWI
 
 | | Fab arm of antibody 10G4 bound to CoV-2 receptor binding domain (RBD) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Heavy chain of Fab arm of antibody 10G4, ... | | Authors: | Langley, D.B, Christ, D. | | Deposit date: | 2022-05-19 | | Release date: | 2023-05-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.873 Å) | | Cite: | Neutralization of CoV-2 omicron lineages by affinity-matured class 5 antibodies
To Be Published
|
|
8CWJ
 
 | |
8CWK
 
 | |
8DXU
 
 | |
7MSQ
 
 | |
5V94
 
 | | Pekin duck egg lysozyme isoform III (DEL-III), cubic form | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PHOSPHATE ION, ... | | Authors: | Langley, D.B, Christ, D. | | Deposit date: | 2017-03-22 | | Release date: | 2017-11-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis of antigen recognition: crystal structure of duck egg lysozyme.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5V8G
 
 | | Pekin duck lysozyme isoform I (DEL-I) | | Descriptor: | CHLORIDE ION, THIOCYANATE ION, lysozyme isoform I | | Authors: | Langley, D.B, Christ, D. | | Deposit date: | 2017-03-22 | | Release date: | 2017-11-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural basis of antigen recognition: crystal structure of duck egg lysozyme.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5V92
 
 | |
7RT9
 
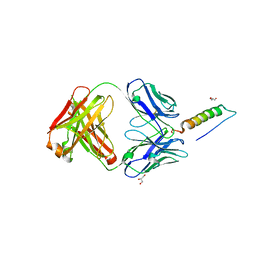 | | Crystal structures of human PYY and NPY | | Descriptor: | 4A3B2-A Fab heavy chain, 4A3B2-A Fab light chain, GLYCEROL, ... | | Authors: | Langley, D.B, Christ, D. | | Deposit date: | 2021-08-12 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of human neuropeptide Y (NPY) and peptide YY (PYY).
Neuropeptides, 92, 2022
|
|
7RTA
 
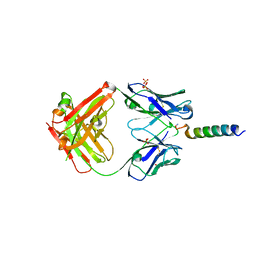 | | Crystal structures of human PYY and NPY | | Descriptor: | 4A3B2-B Fab heavy chain, 4A3B2-B Fab light chain, Neuropeptide Y, ... | | Authors: | Langley, D.B, Christ, D. | | Deposit date: | 2021-08-12 | | Release date: | 2022-03-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of human neuropeptide Y (NPY) and peptide YY (PYY).
Neuropeptides, 92, 2022
|
|
6P4B
 
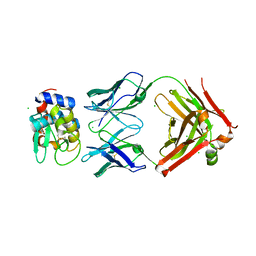 | | HyHEL10 fab variant HyHEL10-4x (heavy chain mutations L4F, Y33H, S56N, and Y58F) bound to hen egg lysozyme variant HEL2x-flex (mutations R21Q, R73E, C76S, and C94S) | | Descriptor: | CHLORIDE ION, HyHEL10 Fab heavy chain, HyHEL10 Fab light chain, ... | | Authors: | Langley, D.B, Christ, D. | | Deposit date: | 2019-05-27 | | Release date: | 2020-05-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conformational diversity facilitates antibody mutation trajectories and discrimination between foreign and self-antigens.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6P4C
 
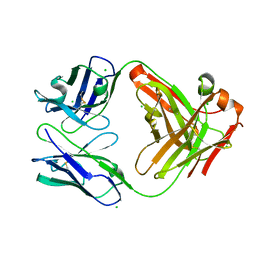 | | HyHEL10 Fab carrying four heavy chain mutations (HyHEL10-4x): L4F, Y33H, S56N, and Y58F | | Descriptor: | CHLORIDE ION, HyHEL10 Fab heavy chain, HyHEL10 Fab light chain | | Authors: | Langley, D.B, Christ, D. | | Deposit date: | 2019-05-27 | | Release date: | 2020-05-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Conformational diversity facilitates antibody mutation trajectories and discrimination between foreign and self-antigens.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6P4D
 
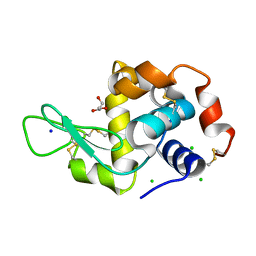 | | Hen egg lysozyme (HEL) containing three point mutations (HEL3x): R21Q, R73E, and D101R | | Descriptor: | CHLORIDE ION, GLYCEROL, Lysozyme C, ... | | Authors: | Langley, D.B, Christ, D. | | Deposit date: | 2019-05-27 | | Release date: | 2020-05-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Conformational diversity facilitates antibody mutation trajectories and discrimination between foreign and self-antigens.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6P4A
 
 | |
5V3B
 
 | |
5V3P
 
 | |
