6NQ7
 
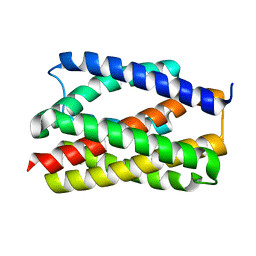 | |
3OS5
 
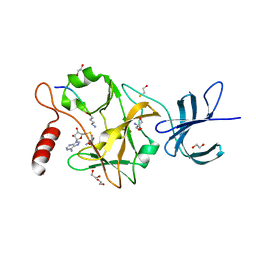 | | SET7/9-Dnmt1 K142me1 complex | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, Dnmt1, ... | | Authors: | Esteve, P.-O, Chang, Y, Samaranayake, M, Upadhyay, A.K, Horton, J.R, Feehery, G.R, Cheng, X, Pradhan, S. | | Deposit date: | 2010-09-08 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | A methylation and phosphorylation switch between an adjacent lysine and serine determines human DNMT1 stability.
Nat.Struct.Mol.Biol., 18, 2011
|
|
2M75
 
 | |
2M7H
 
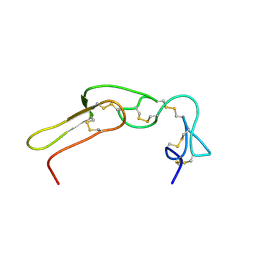 | |
2LJV
 
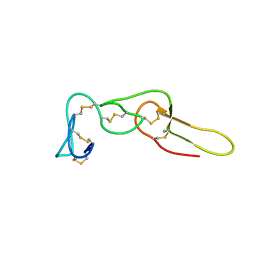 | | Solution structure of Rhodostomin G50L mutant | | Descriptor: | Disintegrin rhodostomin | | Authors: | Chuang, W, Shiu, J, Chen, C, Chen, Y, Chang, Y, Huang, C. | | Deposit date: | 2011-09-29 | | Release date: | 2012-10-03 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Design of Integrin AlphaVbeta3-Specific Disintegrin for Cancer Therapy
To be Published
|
|
2M7F
 
 | |
6XGZ
 
 | | Crystal structure of E. coli MlaFB ABC transport subunits in the monomeric state | | Descriptor: | 1,2-ETHANEDIOL, ABC transporter maintaining OM lipid asymmetry, cytoplasmic STAS component, ... | | Authors: | Chang, Y, Bhabha, G, Ekiert, D.C. | | Deposit date: | 2020-06-18 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of MlaFB uncovers novel mechanisms of ABC transporter regulation.
Elife, 9, 2020
|
|
6XGY
 
 | | Crystal structure of E. coli MlaFB ABC transport subunits in the dimeric state | | Descriptor: | ABC transporter maintaining OM lipid asymmetry, cytoplasmic STAS component, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Chang, Y, Bhabha, G, Ekiert, D.C. | | Deposit date: | 2020-06-18 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of MlaFB uncovers novel mechanisms of ABC transporter regulation.
Elife, 9, 2020
|
|
6WTQ
 
 | | Human JAK2 JH1 domain in complex with PROTAC-intermediate linker handle 4 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, N-methyl-4-{[4-(1-propyl-1H-pyrazol-4-yl)-7H-pyrrolo[2,3-d]pyrimidin-2-yl]amino}benzamide, ... | | Authors: | Yu, S, Nithianantham, S, Fischer, M. | | Deposit date: | 2020-05-03 | | Release date: | 2021-05-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.79968476 Å) | | Cite: | Degradation of Janus kinases in CRLF2-rearranged acute lymphoblastic leukemia.
Blood, 138, 2021
|
|
6WTN
 
 | | Human JAK2 JH1 domain in complex with Ruxolitinib | | Descriptor: | (3R)-3-cyclopentyl-3-[4-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)-1H-pyrazol-1-yl]propanenitrile, 1,2-ETHANEDIOL, Tyrosine-protein kinase JAK2 | | Authors: | Yu, S, Nithianantham, S, Fischer, M. | | Deposit date: | 2020-05-03 | | Release date: | 2021-05-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Degradation of Janus kinases in CRLF2-rearranged acute lymphoblastic leukemia.
Blood, 138, 2021
|
|
6WTP
 
 | | Human JAK2 JH1 domain in complex with PROTAC-intermediate linker handle 3 | | Descriptor: | GLYCEROL, Tyrosine-protein kinase JAK2, tert-butyl 4-[(4-{1-[3-(cyanomethyl)-1-(ethylsulfonyl)azetidin-3-yl]-1H-pyrazol-4-yl}-7H-pyrrolo[2,3-d]pyrimidin-2-yl)amino]benzoate | | Authors: | Yu, S, Nithianantham, S, Fischer, M. | | Deposit date: | 2020-05-03 | | Release date: | 2021-05-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Degradation of Janus kinases in CRLF2-rearranged acute lymphoblastic leukemia.
Blood, 138, 2021
|
|
6WTO
 
 | | Human JAK2 JH1 domain in complex with Baricitinib | | Descriptor: | 1,2-ETHANEDIOL, Baricitinib, Tyrosine-protein kinase JAK2 | | Authors: | Yu, S, Nithianantham, S, Fischer, M. | | Deposit date: | 2020-05-03 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Degradation of Janus kinases in CRLF2-rearranged acute lymphoblastic leukemia.
Blood, 138, 2021
|
|
7UDK
 
 | | Crystal structure of designed helical repeat protein RPB_LRP2_R4 bound to LRPx4 peptide | | Descriptor: | 4xLRP, Designed helical repeat protein (DHR) RPB_LRP2_R4 | | Authors: | Chang, Y, Redler, R.L, Bhabha, G, Ekiert, D.C. | | Deposit date: | 2022-03-20 | | Release date: | 2023-03-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | De novo design of modular peptide-binding proteins by superhelical matching.
Nature, 616, 2023
|
|
7UDL
 
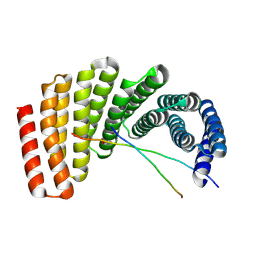 | | Crystal structure of designed helical repeat protein RPB_PLP1_R6 bound to PLPx6 peptide | | Descriptor: | 1,2-ETHANEDIOL, 6xPLP Peptide, Designed helical repeat protein (DHR) RPB_PLP1_R6 | | Authors: | Chang, Y, Redler, R.L, Bhabha, G, Ekiert, D.C. | | Deposit date: | 2022-03-20 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | De novo design of modular peptide-binding proteins by superhelical matching.
Nature, 616, 2023
|
|
7UDN
 
 | |
7UDM
 
 | | Crystal structure of designed helical repeat protein RPB_PLP1_R6 in alternative conformation 1 (with peptide) | | Descriptor: | 6xPLP, Designed helical repeat protein (DHR) RPB_PLP1_R6 | | Authors: | Chang, Y, Redler, R.L, Bhabha, G, Ekiert, D.C. | | Deposit date: | 2022-03-20 | | Release date: | 2023-03-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | De novo design of modular peptide-binding proteins by superhelical matching.
Nature, 616, 2023
|
|
7UE2
 
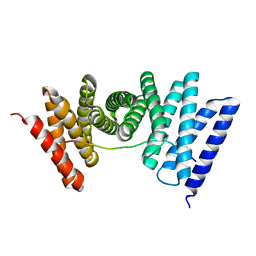 | |
7UPQ
 
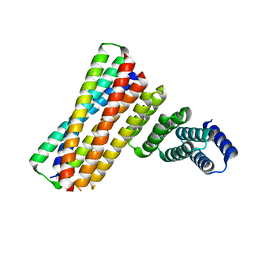 | |
7UPO
 
 | |
7UPP
 
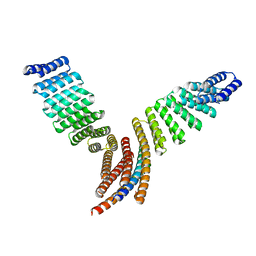 | |
5HPG
 
 | |
5X5O
 
 | | Crystal structure of ZAK in complex with compound D2829 | | Descriptor: | Mitogen-activated protein kinase kinase kinase MLT, N-[2,4-bis(fluoranyl)-3-[2-(3-methoxy-1H-pyrazolo[3,4-b]pyridin-5-yl)ethynyl]phenyl]-3-bromanyl-benzenesulfonamide | | Authors: | Dai, Y.B, Zhao, P, Yun, C.H. | | Deposit date: | 2017-02-17 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.868 Å) | | Cite: | Structure Based Design of N-(3-((1H-Pyrazolo[3,4-b]pyridin-5-yl)ethynyl)benzenesulfonamides as Selective Leucine-Zipper and Sterile-alpha Motif Kinase (ZAK) Inhibitors.
J. Med. Chem., 60, 2017
|
|
8CYG
 
 | |
5JWQ
 
 | | Crystal structure of KaiC S431E in complex with foldswitch-stabilized KaiB from Thermosynechococcus elongatus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Circadian clock protein KaiB, Circadian clock protein kinase KaiC | | Authors: | Tseng, R, Goularte, N.F, Chavan, A, Luu, J, Chang, Y, Heilser, J, Tripathi, S, LiWang, A, Partch, C.L. | | Deposit date: | 2016-05-12 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.871 Å) | | Cite: | Structural basis of the day-night transition in a bacterial circadian clock.
Science, 355, 2017
|
|
5JWO
 
 | | Crystal structure of foldswitch-stabilized KaiB in complex with the N-terminal CI domain of KaiC from Thermosynechococcus elongatus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Circadian clock protein KaiB, Circadian clock protein kinase KaiC | | Authors: | Tseng, R, Goularte, N.F, Chavan, A, Luu, J, Chang, Y, Heilser, J, Tripathi, S, LiWang, A, Partch, C.L. | | Deposit date: | 2016-05-12 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of the day-night transition in a bacterial circadian clock.
Science, 355, 2017
|
|
