8ISO
 
 | | Crystal structure of extended-spectrum class A beta-lactamase, CESS-1 | | Descriptor: | 1,2-ETHANEDIOL, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, Beta-lactamase | | Authors: | Jeong, B.G, Kim, M.Y, Jeong, C.S, Do, H.W, Lee, J.H, Cha, S.S. | | Deposit date: | 2023-03-21 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Characterization of the extended substrate spectrum of the class A beta-lactamase CESS-1 from Stenotrophomonas sp. and structure-based investigation into its substrate preference.
Int J Antimicrob Agents, 63, 2024
|
|
8ISP
 
 | | Crystal structure of extended-spectrum class A beta-lactamase, CESS-1 E166Q acylated by cephalexin | | Descriptor: | (R)-2-((R)-((R)-2-amino-2-phenylacetamido)(carboxy)methyl)-5-methyl-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Beta-lactamase | | Authors: | Jeong, B.G, Kim, M.Y, Jeong, C.S, Do, H.W, Lee, J.H, Cha, S.S. | | Deposit date: | 2023-03-21 | | Release date: | 2024-05-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Characterization of the extended substrate spectrum of the class A beta-lactamase CESS-1 from Stenotrophomonas sp. and structure-based investigation into its substrate preference.
Int J Antimicrob Agents, 63, 2024
|
|
8ISQ
 
 | | Crystal structure of extended-spectrum class A beta-lactamase, CESS-1 E166Q acylated by ampicillin | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-lactamase, ... | | Authors: | Jeong, B.G, Kim, M.Y, Jeong, C.S, Do, H.W, Lee, J.H, Cha, S.S. | | Deposit date: | 2023-03-21 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Characterization of the extended substrate spectrum of the class A beta-lactamase CESS-1 from Stenotrophomonas sp. and structure-based investigation into its substrate preference.
Int J Antimicrob Agents, 63, 2024
|
|
8ISR
 
 | | Crystal structure of extended-spectrum class A beta-lactamase, CESS-1 E166Q acylated by cefaclor | | Descriptor: | (R)-2-((R)-((R)-2-amino-2-phenylacetamido)(carboxy)methyl)-5-chloro-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase | | Authors: | Jeong, B.G, Kim, M.Y, Jeong, C.S, Do, H.W, Lee, J.H, Cha, S.S. | | Deposit date: | 2023-03-21 | | Release date: | 2024-05-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Characterization of the extended substrate spectrum of the class A beta-lactamase CESS-1 from Stenotrophomonas sp. and structure-based investigation into its substrate preference.
Int J Antimicrob Agents, 63, 2024
|
|
6M5H
 
 | |
6M5P
 
 | | A class C beta-lactamase | | Descriptor: | (4R,5S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-4,5-d ihydro-1H-pyrrole-2-carboxylic acid, Beta-lactamase | | Authors: | Bae, D.W, Jung, Y.E, Cha, S.S. | | Deposit date: | 2020-03-11 | | Release date: | 2021-01-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Novel inhibition mechanism of carbapenems on the ACC-1 class C beta-lactamase.
Arch.Biochem.Biophys., 693, 2020
|
|
8KEM
 
 | | PKS domains-fused AmpC EC2 | | Descriptor: | SDR family NAD(P)-dependent oxidoreductase,Beta-lactamase | | Authors: | Son, S.Y, Bae, D.W, Cha, S.S. | | Deposit date: | 2023-08-12 | | Release date: | 2024-06-05 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.76998281 Å) | | Cite: | Structural investigation of the docking domain assembly from trans-AT polyketide synthases.
Structure, 32, 2024
|
|
5K1D
 
 | | Crystal structure of a class C beta lactamase/compound1 complex | | Descriptor: | Beta-lactamase, CADMIUM ION, GUANOSINE-5'-MONOPHOSPHATE | | Authors: | AN, Y.J, Na, J.H, Cha, S.S. | | Deposit date: | 2016-05-18 | | Release date: | 2017-05-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | GMP and IMP Are Competitive Inhibitors of CMY-10, an Extended-Spectrum Class C beta-Lactamase.
Antimicrob. Agents Chemother., 61, 2017
|
|
5K1F
 
 | | Crystal structure of a class C beta lactamase/compound2 complex | | Descriptor: | Beta-lactamase, CADMIUM ION, INOSINIC ACID | | Authors: | An, Y.J, Na, J.H, Cha, S.S. | | Deposit date: | 2016-05-18 | | Release date: | 2017-05-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | GMP and IMP Are Competitive Inhibitors of CMY-10, an Extended-Spectrum Class C beta-Lactamase.
Antimicrob. Agents Chemother., 61, 2017
|
|
5JOC
 
 | | Crystal structure of the S61A mutant of AmpC BER | | Descriptor: | Beta-lactamase, CITRIC ACID | | Authors: | Na, J.H, An, Y.J, Cha, S.S. | | Deposit date: | 2016-05-02 | | Release date: | 2017-05-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for the extended substrate spectrum of AmpC BER and structure-guided discovery of the inhibition activity of citrate against the class C beta-lactamases AmpC BER and CMY-10.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
8HU7
 
 | |
8HUE
 
 | | Crystal structure of FGF2-M2 mutant - D28E/C78I/C96I/S137P | | Descriptor: | 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, Fibroblast growth factor 2 | | Authors: | Jung, Y.E, Cha, S.S, An, Y.J. | | Deposit date: | 2022-12-23 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural and biochemical investigation into stable FGF2 mutants with novel mutation sites and hydrophobic replacements for surface-exposed cysteines.
Plos One, 19, 2024
|
|
7YGK
 
 | |
8HNP
 
 | | Archaeal transcription factor Mutant | | Descriptor: | Archaeal transcription regulator | | Authors: | Bae, D.W, Cha, S.S. | | Deposit date: | 2022-12-08 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.39 Å) | | Cite: | An archaeal transcription factor EnfR with a novel 'eighth note' fold controls hydrogen production of a hyperthermophilic archaeon Thermococcus onnurineus NA1.
Nucleic Acids Res., 51, 2023
|
|
8HNO
 
 | | Archaeal transcription factor Wild type | | Descriptor: | Archaeal transcription regulator | | Authors: | Bae, D.W, Cha, S.S. | | Deposit date: | 2022-12-08 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | An archaeal transcription factor EnfR with a novel 'eighth note' fold controls hydrogen production of a hyperthermophilic archaeon Thermococcus onnurineus NA1.
Nucleic Acids Res., 51, 2023
|
|
8IVU
 
 | | Crystal Structure of Human NAMPT in complex with A4276 | | Descriptor: | N-[[4-(6-methyl-1,3-benzoxazol-2-yl)phenyl]methyl]pyridine-3-carboxamide, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Kang, B.G, Cha, S.S. | | Deposit date: | 2023-03-28 | | Release date: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.09000921 Å) | | Cite: | Discovery of a novel NAMPT inhibitor that selectively targets NAPRT-deficient EMT-subtype cancer cells and alleviates chemotherapy-induced peripheral neuropathy.
Theranostics, 13, 2023
|
|
4ZPX
 
 | |
5F1G
 
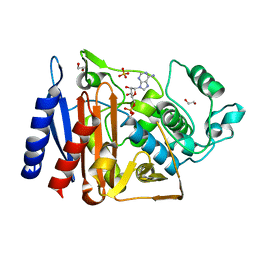 | | Crystal structure of AmpC BER adenylylated in the cytoplasm | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, Beta-lactamase, ... | | Authors: | An, Y.J, Kim, M.K, Na, J.H, Cha, S.S. | | Deposit date: | 2015-11-30 | | Release date: | 2016-12-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural and mechanistic insights into the inhibition of class C beta-lactamases through the adenylylation of the nucleophilic serine.
J.Antimicrob.Chemother., 72, 2017
|
|
5F1F
 
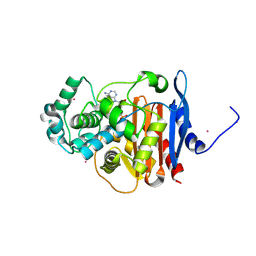 | | Crystal structure of CMY-10 adenylylated by acetyl-AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Beta-lactamase, CADMIUM ION | | Authors: | An, Y.J, Kim, M.K, Na, J.H, Cha, S.S. | | Deposit date: | 2015-11-30 | | Release date: | 2016-12-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.548 Å) | | Cite: | Structural and mechanistic insights into the inhibition of class C beta-lactamases through the adenylylation of the nucleophilic serine.
J.Antimicrob.Chemother., 72, 2017
|
|
7DCK
 
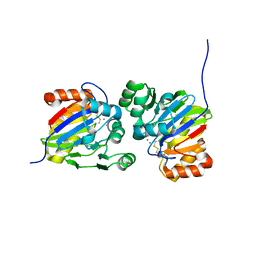 | | Crystal structure of phosphodiesterase tw9814 | | Descriptor: | Lactamase_B domain-containing protein, MANGANESE (II) ION | | Authors: | Heo, Y, Yun, J.H, Park, J.H, Park, S.B, Cha, S.S, Lee, W. | | Deposit date: | 2020-10-26 | | Release date: | 2021-10-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural and functional identification of the uncharacterized metallo-beta-lactamase superfamily protein TW9814 as a phosphodiesterase with unique metal coordination.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7E3Z
 
 | | Non-Ribosomal Peptide Synthetases, Thioesterase | | Descriptor: | CHLORIDE ION, thioesterase | | Authors: | Jung, Y.E, Cha, S.S. | | Deposit date: | 2021-02-09 | | Release date: | 2021-12-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Unprecedented Noncanonical Features of the Nonlinear Nonribosomal Peptide Synthetase Assembly Line for WS9326A Biosynthesis.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7CIN
 
 | |
3L18
 
 | |
5Z8A
 
 | | Crystal structure of GenB1 from Micromonospora echinospora in complex with JI-20A and PLP (external aldimine) | | Descriptor: | (2~{R},3~{R},4~{R},5~{R})-2-[(1~{S},2~{S},3~{R},4~{S},6~{R})-3-[(2~{R},3~{R},4~{R},5~{S},6~{R})-6-(aminomethyl)-3-azany l-4,5-bis(oxidanyl)oxan-2-yl]oxy-4,6-bis(azanyl)-2-oxidanyl-cyclohexyl]oxy-5-methyl-4-(methylamino)oxane-3,5-diol, C-6' aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Hong, S.K, Cha, S.S. | | Deposit date: | 2018-01-31 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.398 Å) | | Cite: | Complete reconstitution of the diverse pathways of gentamicin B biosynthesis.
Nat. Chem. Biol., 15, 2019
|
|
5X9V
 
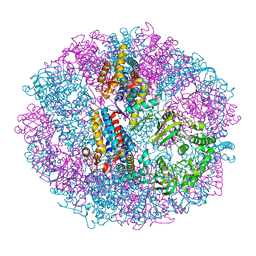 | | Crystal structure of group III chaperonin in the Closed state | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Thermosome, ... | | Authors: | An, Y.J, Cha, S.S. | | Deposit date: | 2017-03-09 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | Structural and mechanistic characterization of an archaeal-like chaperonin from a thermophilic bacterium
Nat Commun, 8, 2017
|
|
