1SGG
 
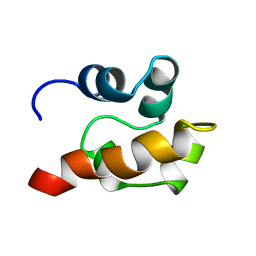 | | THE SOLUTION STRUCTURE OF SAM DOMAIN FROM THE RECEPTOR TYROSINE KINASE EPHB2, NMR, 10 STRUCTURES | | Descriptor: | EPHRIN TYPE-B RECEPTOR 2 | | Authors: | Smalla, M, Schmieder, P, Kelly, M, Ter Laak, A, Krause, G, Ball, L, Wahl, M, Bork, P, Oschkinat, H. | | Deposit date: | 1999-01-08 | | Release date: | 1999-10-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the receptor tyrosine kinase EphB2 SAM domain and identification of two distinct homotypic interaction sites.
Protein Sci., 8, 1999
|
|
4BBQ
 
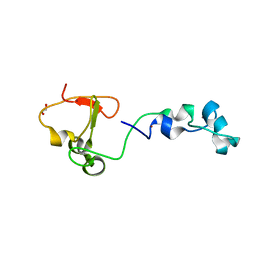 | | Crystal structure of the CXXC and PHD domain of Human Lysine-specific Demethylase 2A (KDM2A)(FBXL11) | | Descriptor: | 1,2-ETHANEDIOL, LYSINE-SPECIFIC DEMETHYLASE 2A, ZINC ION | | Authors: | Allerston, C.K, Watson, A.A, Edlich, C, Li, B, Chen, Y, Ball, L, Krojer, T, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Laue, E.D, Gileadi, O. | | Deposit date: | 2012-09-27 | | Release date: | 2012-10-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Crystal Structure of the Cxxc and Phd Domain of Human Lysine-Specific Demethylase 2A (Kdm2A)(Fbxl11)
To be Published
|
|
2C47
 
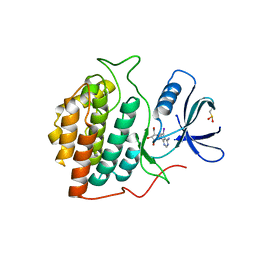 | | Structure of casein kinase 1 gamma 2 | | Descriptor: | (2R,3R,4S,5R)-2-(4-AMINO-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL, CASEIN KINASE 1 GAMMA 2 ISOFORM, MAGNESIUM ION | | Authors: | Bunkoczi, G, Rellos, P, Das, S, Ugochukwu, E, Fedorov, O, Sobott, F, Eswaran, J, Amos, A, Ball, L, von Delft, F, Bullock, A, Debreczeni, J, Turnbull, A, Sundstrom, M, Weigelt, J, Arrowsmith, C, Edwards, A, Knapp, S. | | Deposit date: | 2005-10-16 | | Release date: | 2005-11-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Structure of Casein Kinase 1 Gamma 2
To be Published
|
|
2LG1
 
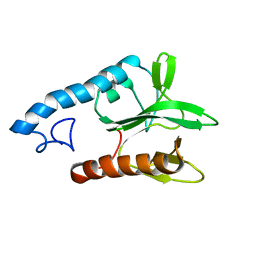 | |
6RN1
 
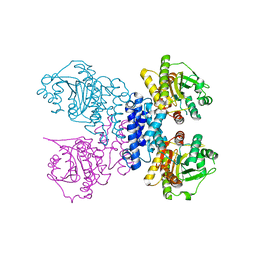 | | Structure of N-terminal truncated Plasmodium falciparum IMP-nucleotidase | | Descriptor: | IMP-specific 5'-nucleotidase, putative | | Authors: | Carrique, L, Ballut, L, Violot, S, Aghajari, N. | | Deposit date: | 2019-05-07 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and catalytic regulation of Plasmodium falciparum IMP specific nucleotidase.
Nat Commun, 11, 2020
|
|
6RNH
 
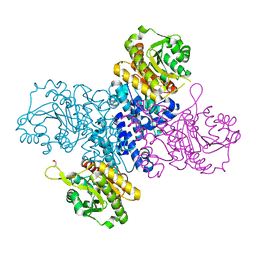 | | Structure of C-terminal truncated Plasmodium falciparum IMP-nucleotidase | | Descriptor: | GLYCEROL, IMP-specific 5'-nucleotidase, putative | | Authors: | Carrique, L, Ballut, L, Violot, S, Aghajari, N. | | Deposit date: | 2019-05-08 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structure and catalytic regulation of Plasmodium falciparum IMP specific nucleotidase.
Nat Commun, 11, 2020
|
|
6RMW
 
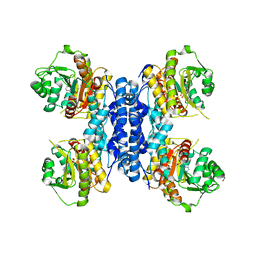 | | Structure of N-terminal truncated IMP bound Plasmodium falciparum IMP-nucleotidase | | Descriptor: | GLYCEROL, IMP-specific 5'-nucleotidase, putative, ... | | Authors: | Carrique, L, Ballut, L, Violot, S, Aghajari, N. | | Deposit date: | 2019-05-07 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure and catalytic regulation of Plasmodium falciparum IMP specific nucleotidase.
Nat Commun, 11, 2020
|
|
6RMO
 
 | | Structure of Plasmodium falciparum IMP-nucleotidase | | Descriptor: | IMP-specific 5'-nucleotidase, putative | | Authors: | Carrique, L, Ballut, L, Violot, S, Aghajari, N. | | Deposit date: | 2019-05-07 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and catalytic regulation of Plasmodium falciparum IMP specific nucleotidase.
Nat Commun, 11, 2020
|
|
7QP1
 
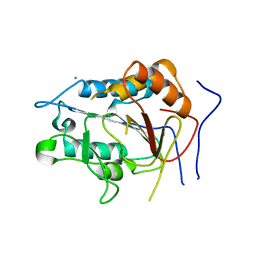 | | Crystal structure of metacaspase from candida glabrata with calcium | | Descriptor: | CALCIUM ION, CHLORIDE ION, Metacaspase-1 | | Authors: | Conchou, L, Ballut, L, Violot, S, Aghajari, N. | | Deposit date: | 2021-12-30 | | Release date: | 2023-01-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and molecular determinants of Candida glabrata metacaspase maturation and activation by calcium.
Commun Biol, 5, 2022
|
|
7QP0
 
 | | Crystal structure of metacaspase from candida glabrata with magnesium | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MAGNESIUM ION, Metacaspase-1 | | Authors: | Conchou, L, Ballut, L, Violot, S, Aghajari, N. | | Deposit date: | 2021-12-30 | | Release date: | 2023-01-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and molecular determinants of Candida glabrata metacaspase maturation and activation by calcium.
Commun Biol, 5, 2022
|
|
6RMD
 
 | | Structure of ATP bound Plasmodium falciparum IMP-nucleotidase | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, IMP-specific 5'-nucleotidase, ... | | Authors: | Carrique, L, Ballut, L, Violot, S, Aghajari, N. | | Deposit date: | 2019-05-06 | | Release date: | 2020-07-15 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and catalytic regulation of Plasmodium falciparum IMP specific nucleotidase.
Nat Commun, 11, 2020
|
|
6RME
 
 | | Structure of IMP bound Plasmodium falciparum IMP-nucleotidase mutant D172N | | Descriptor: | GLYCEROL, IMP-specific 5'-nucleotidase, putative, ... | | Authors: | Carrique, L, Ballut, L, Violot, S, Aghajari, N. | | Deposit date: | 2019-05-06 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure and catalytic regulation of Plasmodium falciparum IMP specific nucleotidase.
Nat Commun, 11, 2020
|
|
1EGX
 
 | | SOLUTION STRUCTURE OF THE ENA-VASP HOMOLOGY 1 (EVH1) DOMAIN OF HUMAN VASODILATOR-STIMULATED PHOSPHOPROTEIN (VASP) | | Descriptor: | VASODILATOR-STIMULATED PHOSPHOPROTEIN | | Authors: | Ball, L, Kuhne, R, Hoffmann, B, Hafner, A, Schmieder, P, Volkmer-Engert, R, Hof, M, Wahl, M, Schneider-Mergener, J, Walter, U, Oschkinat, H, Jarchau, T. | | Deposit date: | 2000-02-17 | | Release date: | 2000-09-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Dual epitope recognition by the VASP EVH1 domain modulates polyproline ligand specificity and binding affinity.
EMBO J., 19, 2000
|
|
2ES0
 
 | | Structure of the regulator of G-protein signaling domain of RGS6 | | Descriptor: | regulator of G-protein signalling 6 | | Authors: | Schoch, G.A, Phillips, C, Turnbull, A, Niesen, F, Johansson, C, Elkins, J.M, Longman, E, Gilealdi, C, Sobott, F, Ball, L, Sundstrom, M, Edwards, A, Arrowsmith, C, von Delft, F, Doyle, D.A, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-10-25 | | Release date: | 2005-11-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural diversity in the RGS domain and its interaction with heterotrimeric G protein alpha-subunits.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
1ZV4
 
 | | Structure of the Regulator of G-Protein Signaling 17 (RGSZ2) | | Descriptor: | Regulator of G-protein signaling 17 | | Authors: | Schoch, G.A, Jansson, A, Elkins, J.M, Haroniti, A, Niesen, F.H, Bunkoczi, G, Lee, W.H, Turnbull, A.P, Yang, X, Sundstrom, M, Arrowsmith, C, Edwards, A, Marsden, B, Gileadi, O, Ball, L, von Delft, F, Doyle, D.A, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-06-01 | | Release date: | 2005-06-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural diversity in the RGS domain and its interaction with heterotrimeric G protein alpha-subunits.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
1Z57
 
 | | Crystal structure of human CLK1 in complex with 10Z-Hymenialdisine | | Descriptor: | DEBROMOHYMENIALDISINE, Dual specificity protein kinase CLK1 | | Authors: | Debreczeni, J, Das, S, Knapp, S, Bullock, A, Guo, K, Amos, A, Fedorov, O, Edwards, A, Sundstrom, M, von Delft, F, Niesen, F.H, Ball, L, Sobott, F, Arrowsmith, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-03-17 | | Release date: | 2005-04-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Kinase domain insertions define distinct roles of CLK kinases in SR protein phosphorylation.
Structure, 17, 2009
|
|
2AHS
 
 | | Crystal Structure of the Catalytic Domain of Human Tyrosine Receptor Phosphatase Beta | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Receptor-type tyrosine-protein phosphatase beta, ... | | Authors: | Ugochukwu, E, Eswaran, J, Barr, A, Gileadi, O, Sobott, F, Burgess, N, Ball, L, Bray, J, von Delft, F, Debreczeni, J, Bunkoczi, G, Turnbull, A, Das, S, Weigelt, J, Edwards, A, Arrowsmith, C, Sundstrom, M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-07-28 | | Release date: | 2005-08-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Large-scale structural analysis of the classical human protein tyrosine phosphatome.
Cell(Cambridge,Mass.), 136, 2009
|
|
5K4Z
 
 | | M. thermoresistible IMPDH in complex with IMP and Compound 6 | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase, ~{N}-(4-fluorophenyl)-4-(2~{H}-indazol-6-ylsulfamoyl)-3,5-dimethyl-1~{H}-pyrrole-2-carboxamide | | Authors: | Pacitto, A, Ascher, D.B, Blundell, T.L. | | Deposit date: | 2016-05-22 | | Release date: | 2016-10-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Essential but Not Vulnerable: Indazole Sulfonamides Targeting Inosine Monophosphate Dehydrogenase as Potential Leads against Mycobacterium tuberculosis.
ACS Infect Dis, 3, 2017
|
|
5K4X
 
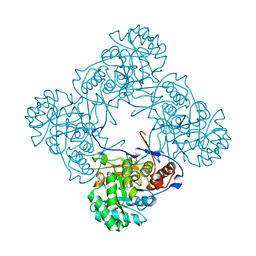 | | M. thermoresistible IMPDH in complex with IMP and Compound 1 | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase, ~{N}-(2~{H}-indazol-6-yl)-3,5-dimethyl-1~{H}-pyrazole-4-sulfonamide | | Authors: | Pacitto, A, Ascher, D.B, Blundell, T.L. | | Deposit date: | 2016-05-22 | | Release date: | 2016-10-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Essential but Not Vulnerable: Indazole Sulfonamides Targeting Inosine Monophosphate Dehydrogenase as Potential Leads against Mycobacterium tuberculosis.
ACS Infect Dis, 3, 2017
|
|
5DU8
 
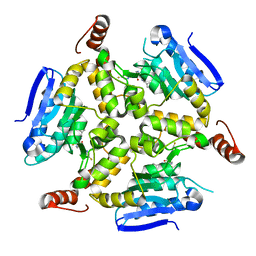 | | Crystal structure of M. tuberculosis EchA6 bound to GSK572A | | Descriptor: | (5R,7S)-5-(4-ethylphenyl)-N-[(5-fluoropyridin-2-yl)methyl]-7-(trifluoromethyl)-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrimidine-3-carboxamide, Probable enoyl-CoA hydratase echA6 | | Authors: | Cox, J.A.G, Besra, G.S, Futterer, K. | | Deposit date: | 2015-09-18 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | THPP target assignment reveals EchA6 as an essential fatty acid shuttle in mycobacteria.
Nat Microbiol, 1, 2016
|
|
5DTP
 
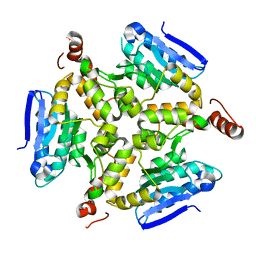 | |
5DU4
 
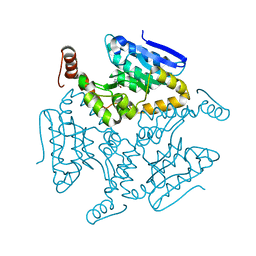 | | Crystal structure of M. tuberculosis EchA6 bound to ligand GSK366A | | Descriptor: | (5R,7S)-5-(4-ethylphenyl)-N-(4-methoxybenzyl)-7-(trifluoromethyl)-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrimidine-3-carboxamide, Probable enoyl-CoA hydratase echA6 | | Authors: | Cox, J.A.G, Besra, G.S, Futterer, K. | | Deposit date: | 2015-09-18 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | THPP target assignment reveals EchA6 as an essential fatty acid shuttle in mycobacteria.
Nat Microbiol, 1, 2016
|
|
5JFO
 
 | | Structure of the M.tuberculosis enoyl-reductase InhA in complex with GSK625 | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADH], N-{1-[(2-chloro-6-fluorophenyl)methyl]-1H-pyrazol-3-yl}-5-[(1S)-1-(3-methyl-1H-pyrazol-1-yl)ethyl]-1,3,4-thiadiazol-2-amine, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Gulten, G, Sacchettini, J.C. | | Deposit date: | 2016-04-19 | | Release date: | 2016-08-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.907 Å) | | Cite: | Antitubercular drugs for an old target: GSK693 as a promising InhA direct inhibitor.
Ebiomedicine, 8, 2016
|
|
5DU6
 
 | | Crystal structure of M. tuberculosis EchA6 bound to ligand GSK059A. | | Descriptor: | (5R,7R)-5-(4-ethylphenyl)-N-(4-fluorobenzyl)-7-methyl-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrimidine-3-carboxamide, Probable enoyl-CoA hydratase echA6 | | Authors: | Cox, J.A.G, Besra, G.S, Futterer, K. | | Deposit date: | 2015-09-18 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | THPP target assignment reveals EchA6 as an essential fatty acid shuttle in mycobacteria.
Nat Microbiol, 1, 2016
|
|
5MVR
 
 | | Crystal structure of Bacillus subtilus YdiB | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Jault, J.-M, Aghajari, N. | | Deposit date: | 2017-01-17 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.762 Å) | | Cite: | Expanding the Kinome World: A New Protein Kinase Family Widely Conserved in Bacteria.
J. Mol. Biol., 429, 2017
|
|
