8VC8
 
 | |
6O0I
 
 | | NMR ensemble of computationally designed protein XAA | | Descriptor: | Design construct XAA | | Authors: | Wei, K.Y, Moschidi, D, Nerli, S, Sgourakis, N, Baker, D. | | Deposit date: | 2019-02-16 | | Release date: | 2020-04-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Computational design of closely related proteins that adopt two well-defined but structurally divergent folds.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7S5B
 
 | |
6UFA
 
 | | S4 symmetric peptide design number 1, Tim zinc-bound form | | Descriptor: | S4-1, Tim, Zinc-bound form, ... | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-24 | | Release date: | 2020-12-02 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (0.77 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6UG2
 
 | | C2 symmetric peptide design number 1, Zappy, crystal form 2 | | Descriptor: | C2-1, Zappy, crystal form 2, ... | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-25 | | Release date: | 2020-12-02 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6UF4
 
 | | S2 symmetric peptide design number 4 crystal form 2, Pugsley | | Descriptor: | S2-4, Pusgley crystal form 2 | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-23 | | Release date: | 2020-12-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6UF9
 
 | | S4 symmetric peptide design number 1, Tim apo form | | Descriptor: | S4-1, Tim apo-form, SULFATE ION | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-24 | | Release date: | 2020-12-02 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6UG6
 
 | | C3 symmetric peptide design number 1, Sporty, crystal form 2 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, C3-1, ... | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-25 | | Release date: | 2020-12-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6UDW
 
 | | S2 symmetric peptide design number 3 crystal form 2, Lurch | | Descriptor: | S2-3, Lurch crystal form 2 | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-19 | | Release date: | 2020-09-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6UGB
 
 | | C3 symmetric peptide design number 2, Baby Basil | | Descriptor: | C3 symmetric peptide design number 2, Baby Basil, CHLORIDE ION, ... | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-26 | | Release date: | 2020-12-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
8UZL
 
 | | Designed Transmembrane beta-barrel- TMB10_163 | | Descriptor: | Designed Transmembrane beta-barrel TMB10_163, HEXANE-1,6-DIOL | | Authors: | Bera, A.K, Lemma, S.B, Kang, A, Baker, D. | | Deposit date: | 2023-11-15 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Sculpting conducting nanopore size and shape through de novo protein design.
Science, 385, 2024
|
|
8VD6
 
 | |
8VFQ
 
 | | De novo design apixaban-binding protein: apx1049 | | Descriptor: | 1-(4-METHOXYPHENYL)-7-OXO-6-[4-(2-OXOPIPERIDIN-1-YL)PHENYL]-4,5,6,7-TETRAHYDRO-1H-PYRAZOLO[3,4-C]PYRIDINE-3-CARBOXAMIDE, De novo designed apixaban-binding protein apx1049 | | Authors: | Bera, A.K, Lee, G.R, Baker, D. | | Deposit date: | 2023-12-21 | | Release date: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Small-molecule binding and sensing with a designed protein family
To Be Published
|
|
8VEZ
 
 | | De novo design apixaban-binding protein: apx1049 | | Descriptor: | 1-(4-METHOXYPHENYL)-7-OXO-6-[4-(2-OXOPIPERIDIN-1-YL)PHENYL]-4,5,6,7-TETRAHYDRO-1H-PYRAZOLO[3,4-C]PYRIDINE-3-CARBOXAMIDE, De novo design apixaban-binding protein: apx1049 | | Authors: | Bera, A.K, Lee, G.R, Baker, D. | | Deposit date: | 2023-12-20 | | Release date: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Small-molecule binding and sensing with a designed protein family
To Be Published
|
|
9BK6
 
 | | Structure of B10_CYTX binder-complex | | Descriptor: | B10_CYTX binder, Cytotoxin 1 | | Authors: | Bera, A.K, Torres, S.V, Kang, A, Baker, D. | | Deposit date: | 2024-04-26 | | Release date: | 2024-10-09 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | De novo designed proteins neutralize lethal snake venom toxins.
Nature, 639, 2025
|
|
9BK7
 
 | |
9BK5
 
 | | Structure of LNG binder complex | | Descriptor: | Alpha-cobratoxin, LNG binder | | Authors: | Bera, A.K, Torres, S.V, Kang, A, Baker, D. | | Deposit date: | 2024-04-26 | | Release date: | 2024-10-09 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | De novo designed proteins neutralize lethal snake venom toxins.
Nature, 639, 2025
|
|
7FB8
 
 | | De Novo-Designed and Disulfide-Bridged Peptide Heterodimer - hd1 | | Descriptor: | ASP-ASP-LYS-ASP-CYS-ASP-GLU-TYR-CYS-LYS-LYS-THR-LYS-GLU-NH2, GLU-LE1-THR-GLY-HIS-ILE-GLU-GLY-PRO-THR-LE1-THR-LE1-HIS-CYS-LYS-NH2 | | Authors: | Yao, H, Yao, S, Zheng, Y, Moyer, A, Baker, D, Wu, C. | | Deposit date: | 2021-07-08 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | De novo design and directed folding of disulfide-bridged peptide heterodimers.
Nat Commun, 13, 2022
|
|
7FBA
 
 | | De Novo-Designed and Disulfide-Bridged Peptide Heterodimer - hd2 | | Descriptor: | ALA-LE1-CYS-GLU-CYS-GLY-PRO-THR-ARG-GLU-CYS-LYS-NH2, GLU-CYS-ARG-GLU-TYR-GLY-PRO-LE1-LYS-LE1-LE1-ALA-NH2 | | Authors: | Yao, H, Yao, S, Moyer, A, Zheng, Y, Baker, D, Wu, C. | | Deposit date: | 2021-07-08 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | De novo design and directed folding of disulfide-bridged peptide heterodimers.
Nat Commun, 13, 2022
|
|
9CC5
 
 | |
9CC6
 
 | |
9CU8
 
 | | Crystal Structure of TNFR2 TNFR2_mb1 Complex | | Descriptor: | TNFR2_mb1, Tumor necrosis factor receptor superfamily member 1B | | Authors: | Bera, A.K, Glogl, M, Baker, D. | | Deposit date: | 2024-07-26 | | Release date: | 2024-12-04 | | Last modified: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Target-conditioned diffusion generates potent TNFR superfamily antagonists and agonists.
Science, 386, 2024
|
|
6DMP
 
 | | De Novo Design of a Protein Heterodimer with Specificity Mediated by Hydrogen Bond Networks | | Descriptor: | Designed orthogonal protein DHD13_XAAA_A, Designed orthogonal protein DHD13_XAAA_B | | Authors: | Chen, Z, Flores-Solis, D, Sgourakis, N.G, Baker, D. | | Deposit date: | 2018-06-05 | | Release date: | 2018-12-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Programmable design of orthogonal protein heterodimers.
Nature, 565, 2019
|
|
2XYZ
 
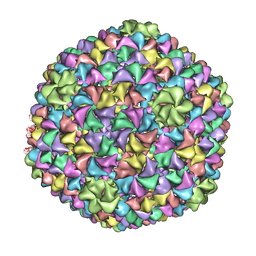 | | De Novo model of Bacteriophage P22 virion coat protein | | Descriptor: | COAT PROTEIN | | Authors: | Chen, D.-H, Baker, M.L, Hryc, C.F, DiMaio, F, Jakana, J, Wu, W, Dougherty, M, Haase-Pettingell, C, Schmid, M.F, Jiang, W, Baker, D, King, J.A, Chiu, W. | | Deposit date: | 2010-11-19 | | Release date: | 2011-02-02 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural Basis for Scaffolding-Mediated Assembly and Maturation of a DsDNA Virus.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2XYY
 
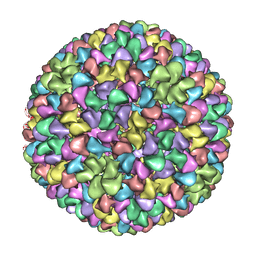 | | De Novo model of Bacteriophage P22 procapsid coat protein | | Descriptor: | COAT PROTEIN | | Authors: | Chen, D.-H, Baker, M.L, Hryc, C.F, DiMaio, F, Jakana, J, Wu, W, Dougherty, M, Haase-Pettingell, C, Schmid, M.F, Jiang, W, Baker, D, King, J.A, Chiu, W. | | Deposit date: | 2010-11-19 | | Release date: | 2011-02-02 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural Basis for Scaffolding-Mediated Assembly and Maturation of a DsDNA Virus.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
