8QAB
 
 | | X-ray crystal structure of a de novo designed antiparallel coiled-coil hexameric alpha-helical barrel with 4 heptad repeats, apCCHex | | Descriptor: | 1-methoxy-2-[2-[2-[2-[2-[2-[2-[2-[2-[2-(2-methoxyethoxy)ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethane, apCCHex | | Authors: | Naudin, E.N, Albanese, K.I, Petrenas, R, Woolfson, D.N. | | Deposit date: | 2023-08-22 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Rationally seeded computational protein design of ɑ-helical barrels.
Nat.Chem.Biol., 20, 2024
|
|
8QAD
 
 | |
8QAI
 
 | |
8QKD
 
 | |
7KH7
 
 | | Crystal structure of Staphylococcus aureus ketol-acid reductoisomerase in complex with Mg2+, NADPH, and NSC116565 | | Descriptor: | 6-hydroxy-2-methyl[1,3]thiazolo[4,5-d]pyrimidine-5,7(4H,6H)-dione, Ketol-acid reductoisomerase (NADP(+)), MAGNESIUM ION, ... | | Authors: | Kurz, J.L, Patel, K.P, Guddat, L.W. | | Deposit date: | 2020-10-20 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Discovery of a Pyrimidinedione Derivative with Potent Inhibitory Activity against Mycobacterium tuberculosis Ketol-Acid Reductoisomerase.
Chemistry, 27, 2021
|
|
7KE2
 
 | | Crystal structure of Staphylococcus aureus ketol-acid reductoisomerase in complex with Mg2+ and NSC116565 | | Descriptor: | 6-hydroxy-2-methyl[1,3]thiazolo[4,5-d]pyrimidine-5,7(4H,6H)-dione, Ketol-acid reductoisomerase (NADP(+)), MAGNESIUM ION | | Authors: | Kurz, J.L, Patel, K.P, Guddat, L.W. | | Deposit date: | 2020-10-09 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Discovery of a Pyrimidinedione Derivative with Potent Inhibitory Activity against Mycobacterium tuberculosis Ketol-Acid Reductoisomerase.
Chemistry, 27, 2021
|
|
6IEQ
 
 | | Crystal Structure of HIV-1 Env ConM SOSIP.v7 in Complex with bNAb PGT124 and 35O22 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 35O22 Fab Heavy Chain, ... | | Authors: | Han, B.W, Wilson, I.A. | | Deposit date: | 2018-09-16 | | Release date: | 2019-05-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structure and immunogenicity of a stabilized HIV-1 envelope trimer based on a group-M consensus sequence.
Nat Commun, 10, 2019
|
|
8HUI
 
 | | Crystal structure of DFA I-forming Inulin Lyase from Streptomyces peucetius subsp. caesius ATCC 27952 in complex with GF4, DFA I, and fructose | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Cheng, M, Rao, Y.J, Mu, W.M. | | Deposit date: | 2022-12-24 | | Release date: | 2023-12-27 | | Last modified: | 2025-01-08 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Structural Insights into the Catalytic Cycle of Inulin Fructotransferase: From Substrate Anchoring to Product Releasing.
J.Agric.Food Chem., 72, 2024
|
|
2ZEB
 
 | | Potent, Nonpeptide Inhibitors of Human Mast Cell Tryptase | | Descriptor: | 1-(1'-{[3-(methylsulfanyl)-2-benzothiophen-1-yl]carbonyl}spiro[1-benzofuran-3,4'-piperidin]-5-yl)methanamine, Tryptase beta 2 | | Authors: | Spurlino, J.C, Lewandowski, F, Milligan, C. | | Deposit date: | 2007-12-08 | | Release date: | 2008-12-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Potent, nonpeptide inhibitors of human mast cell tryptase. Synthesis and biological evaluation of novel spirocyclic piperidine amide derivatives
Bioorg.Med.Chem.Lett., 18, 2008
|
|
8HSN
 
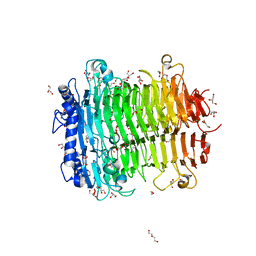 | | Crystal structure of DFA I-forming Inulin Lyase from Streptomyces peucetius subsp. caesius ATCC 27952 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Fructotransferase, ... | | Authors: | Cheng, M, Rao, Y.J, Mu, W.M. | | Deposit date: | 2022-12-20 | | Release date: | 2023-12-27 | | Last modified: | 2025-01-08 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural Insights into the Catalytic Cycle of Inulin Fructotransferase: From Substrate Anchoring to Product Releasing.
J.Agric.Food Chem., 72, 2024
|
|
4X49
 
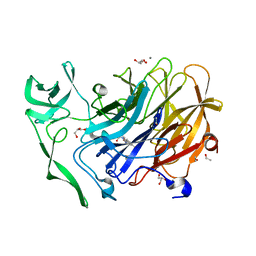 | | Crystal structure of the intramolecular trans-sialidase from Ruminococcus gnavus in complex with oseltamivir carboxylate | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, ACETYL GROUP, Anhydrosialidase, ... | | Authors: | Owen, C.D, Tailford, L.E, Taylor, G.L, Juge, N. | | Deposit date: | 2014-12-02 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Discovery of intramolecular trans-sialidases in human gut microbiota suggests novel mechanisms of mucosal adaptation.
Nat Commun, 6, 2015
|
|
3NXE
 
 | |
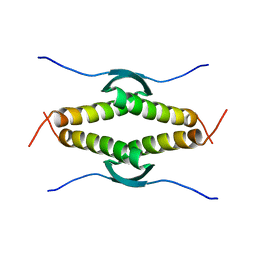
3SAK
 
 | |
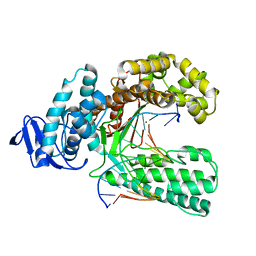
4XIU
 
 | |
6H9X
 
 | | Klebsiella pneumoniae Seryl-tRNA Synthetase in Complex with the Intermediate Analog 5'-O-(N-(L-Seryl)-Sulfamoyl)Adenosine | | Descriptor: | 1,2-ETHANEDIOL, 5'-O-(N-(L-SERYL)-SULFAMOYL)ADENOSINE, CALCIUM ION, ... | | Authors: | Pang, L, De Graef, S, Strelkov, S.V, Weeks, S.D. | | Deposit date: | 2018-08-06 | | Release date: | 2019-05-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Acylated sulfonamide adenosines as potent inhibitors of the adenylate-forming enzyme superfamily.
Eur.J.Med.Chem., 174, 2019
|
|
3BD1
 
 | | Structure of the Cro protein from putative prophage element Xfaso 1 in Xylella fastidiosa strain Ann-1 | | Descriptor: | CHLORIDE ION, Cro protein, GLYCEROL, ... | | Authors: | Hall, B.M, Roberts, S.A, Montfort, W.R, Cordes, M.H. | | Deposit date: | 2007-11-13 | | Release date: | 2008-03-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Transitive homology-guided structural studies lead to discovery of
Cro proteins with 40% sequence identity but different folds
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
4X4A
 
 | | Crystal structure of the intramolecular trans-sialidase from Ruminococcus gnavus in complex with 2,7-Anhydro-Neu5Ac | | Descriptor: | 2-ACETYLAMINO-7-(1,2-DIHYDROXY-ETHYL)-3-HYDROXY-6,8-DIOXA-BICYCLO[3.2.1]OCTANE-5-CARBOXYLIC ACID, ACETYL GROUP, Anhydrosialidase, ... | | Authors: | Owen, C.D, Tailford, L.E, Taylor, G.L, Juge, N. | | Deposit date: | 2014-12-02 | | Release date: | 2015-07-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Discovery of intramolecular trans-sialidases in human gut microbiota suggests novel mechanisms of mucosal adaptation.
Nat Commun, 6, 2015
|
|
4X6K
 
 | | Crystal structure of the intramolecular trans-sialidase from Ruminococcus gnavus in complex with Siastatin B | | Descriptor: | (2S,3R,4S,5S)-2-(acetylamino)-5-carboxy-3,4-dihydroxypiperidinium, ACETYL GROUP, Anhydrosialidase | | Authors: | Owen, C.D, Tailford, L.E, Taylor, G.L, Juge, N. | | Deposit date: | 2014-12-08 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Discovery of intramolecular trans-sialidases in human gut microbiota suggests novel mechanisms of mucosal adaptation.
Nat Commun, 6, 2015
|
|
3NXN
 
 | | X-ray structure of ester chemical analogue 'covalent dimer' [Ile50,O-Ile50']HIV-1 protease complexed with KVS-1 inhibitor | | Descriptor: | N~2~-[(2R,5S)-5-({(2S,3S)-2-[(N-acetyl-L-threonyl)amino]-3-methylpent-4-enoyl}amino)-2-butyl-4,4-dihydroxynonanoyl]-L-glutaminyl-L-argininamide, SULFATE ION, protease covalent dimer | | Authors: | Torbeev, V.Y, Kent, S.B.H. | | Deposit date: | 2010-07-14 | | Release date: | 2011-11-02 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Protein conformational dynamics in the mechanism of HIV-1 protease catalysis.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3NWX
 
 | | X-ray structure of ester chemical analogue [O-Ile50,O-Ile50']HIV-1 protease complexed with KVS-1 inhibitor | | Descriptor: | N~2~-[(2R,5S)-5-({(2S,3S)-2-[(N-acetyl-L-threonyl)amino]-3-methylpent-4-enoyl}amino)-2-butyl-4,4-dihydroxynonanoyl]-L-glutaminyl-L-argininamide, SULFATE ION, protease | | Authors: | Torbeev, V.Y, Kent, S.B.H. | | Deposit date: | 2010-07-12 | | Release date: | 2011-11-02 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Protein conformational dynamics in the mechanism of HIV-1 protease catalysis.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3NYG
 
 | |
3O6A
 
 | | F144Y/F258Y Double Mutant of Exo-beta-1,3-glucanase from Candida albicans at 2 A | | Descriptor: | Glucan 1,3-beta-glucosidase | | Authors: | Nakatani, Y, Cutfield, S.M, Cutfield, J.F. | | Deposit date: | 2010-07-28 | | Release date: | 2010-09-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Carbohydrate binding sites in Candida albicans exo-beta-1,3-glucanase and the role of the Phe-Phe 'clamp' at the active site entrance
Febs J., 277, 2010
|
|
4YP8
 
 | | Irak4-inhibitor co-structure | | Descriptor: | Interleukin-1 receptor-associated kinase 4, N-{1-(4-cyclopropyl-2-fluorophenyl)-3-[1-(propan-2-yl)piperidin-4-yl]-1H-pyrazol-5-yl}pyrazolo[1,5-a]pyrimidine-3-carboxamide | | Authors: | Fischmann, T.O. | | Deposit date: | 2015-03-12 | | Release date: | 2015-05-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.641 Å) | | Cite: | Potent and Selective Amidopyrazole Inhibitors of IRAK4 That Are Efficacious in a Rodent Model of Inflammation.
Acs Med.Chem.Lett., 6, 2015
|
|
4YO6
 
 | | Irak4-inhibitor co-structure | | Descriptor: | Interleukin-1 receptor-associated kinase 4, N-(3-methyl-1-phenyl-1H-pyrazol-5-yl)pyrazolo[1,5-a]pyrimidine-3-carboxamide | | Authors: | Fischmann, T.O. | | Deposit date: | 2015-03-11 | | Release date: | 2015-05-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Potent and Selective Amidopyrazole Inhibitors of IRAK4 That Are Efficacious in a Rodent Model of Inflammation.
Acs Med.Chem.Lett., 6, 2015
|
|
4X47
 
 | | Crystal structure of the intramolecular trans-sialidase from Ruminococcus gnavus in complex with Neu5Ac2en | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, Anhydrosialidase, PHOSPHATE ION | | Authors: | Owen, C.D, Tailford, L.E, Taylor, G.L, Juge, N. | | Deposit date: | 2014-12-02 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of intramolecular trans-sialidases in human gut microbiota suggests novel mechanisms of mucosal adaptation.
Nat Commun, 6, 2015
|
|
