3EMP
 
 | |
5TGL
 
 | | A MODEL FOR INTERFACIAL ACTIVATION IN LIPASES FROM THE STRUCTURE OF A FUNGAL LIPASE-INHIBITOR COMPLEX | | Descriptor: | LIPASE, N-HEXYLPHOSPHONATE ETHYL ESTER | | Authors: | Brzozowski, A.M, Derewenda, U, Derewenda, Z.S, Dodson, G.G, Lawson, D, Turkenburg, J.P, Bjorkling, F, Huge-Jensen, B, Patkar, S.R, Thim, L. | | Deposit date: | 1991-10-30 | | Release date: | 1994-01-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A model for interfacial activation in lipases from the structure of a fungal lipase-inhibitor complex.
Nature, 351, 1991
|
|
3DJ5
 
 | | Crystal structure of the mouse Aurora-A catalytic domain (Asn186->Gly, Lys240->Arg, Met302->Leu) in complex with Compound 290. | | Descriptor: | 3-({3-[(6-amino-5-bromopyrimidin-4-yl)sulfanyl]propanoyl}amino)-4-methoxy-N-phenylbenzamide, serine/threonine kinase 6 | | Authors: | Elling, R.A, Erlanson, D.A, Yang, W, Tangonan, B.T, Hansen, S.K, Romanowski, M.J. | | Deposit date: | 2008-06-22 | | Release date: | 2009-05-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | New fragment-based drug discovery
To be Published
|
|
1FV9
 
 | |
1ZNJ
 
 | | INSULIN, MONOCLINIC CRYSTAL FORM | | Descriptor: | CHLORIDE ION, INSULIN, PHENOL, ... | | Authors: | Turkenburg, M.G.W, Whittingham, J.L, Turkenburg, J.P, Dodson, G.G, Derewenda, U, Smith, G.D, Dodson, E.J, Derewenda, Z.S, Xiao, B. | | Deposit date: | 1997-09-23 | | Release date: | 1998-01-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure Determination and Refinement of Two Crystal Forms of Native Insulins
To be Published
|
|
4MAB
 
 | | Resolving Cys to Ala variant of Salmonella Alkyl Hydroperoxide Reductase C in its substrate-ready conformation | | Descriptor: | Alkyl hydroperoxide reductase subunit C, CHLORIDE ION, GLYCEROL, ... | | Authors: | Perkins, A, Nelson, K.J, Williams, J.R, Poole, L.B, Karplus, P.A. | | Deposit date: | 2013-08-15 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The sensitive balance between the fully folded and locally unfolded conformations of a model peroxiredoxin.
Biochemistry, 52, 2013
|
|
1EV3
 
 | | Structure of the rhombohedral form of the M-cresol/insulin R6 hexamer | | Descriptor: | CHLORIDE ION, INSULIN, M-CRESOL, ... | | Authors: | Smith, G.D, Ciszak, E, Magrum, L.A, Pangborn, W.A, Blessing, R.H. | | Deposit date: | 2000-04-19 | | Release date: | 2000-12-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | R6 hexameric insulin complexed with m-cresol or resorcinol.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
3MB6
 
 | | Human CK2 catalytic domain in complex with a difurane derivative inhibitor (CPA) | | Descriptor: | Casein kinase II subunit alpha, SULFATE ION, naphtho[2,1-b:7,6-b']difuran-2,8-dicarboxylic acid | | Authors: | Reiser, J.-B, Prudent, R, Claude, C. | | Deposit date: | 2010-03-25 | | Release date: | 2010-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | New potent dual inhibitors of CK2 and Pim kinases: discovery and structural insights.
Faseb J., 24, 2010
|
|
1ETG
 
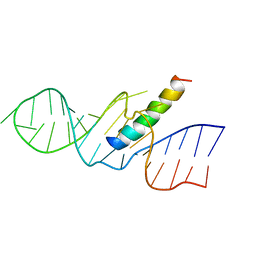 | | REV RESPONSE ELEMENT (RRE) RNA COMPLEXED WITH REV PEPTIDE, NMR, 19 STRUCTURES | | Descriptor: | REV PEPTIDE, REV RESPONSIVE ELEMENT RNA | | Authors: | Battiste, J.L, Mao, H, Rao, N.S, Tan, R, Muhandiram, D.R, Kay, L.E, Frankel, A.D, Willamson, J.R. | | Deposit date: | 1996-08-28 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Alpha helix-RNA major groove recognition in an HIV-1 rev peptide-RRE RNA complex.
Science, 273, 1996
|
|
1ETF
 
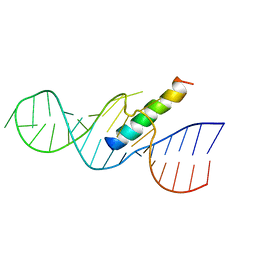 | | REV RESPONSE ELEMENT (RRE) RNA COMPLEXED WITH REV PEPTIDE, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | REV PEPTIDE, REV RESPONSE ELEMENT RNA | | Authors: | Battiste, J.L, Mao, H, Rao, N.S, Tan, R, Muhandiram, D.R, Kay, L.E, Frankel, A.D, Willamson, J.R. | | Deposit date: | 1996-08-28 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Alpha helix-RNA major groove recognition in an HIV-1 rev peptide-RRE RNA complex.
Science, 273, 1996
|
|
1FIH
 
 | | N-ACETYLGALACTOSAMINE BINDING MUTANT OF MANNOSE-BINDING PROTEIN A (QPDWG-HDRPY), COMPLEX WITH N-ACETYLGALACTOSAMINE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Feinberg, H, Torgerson, D, Drickamer, K, Weis, W.I. | | Deposit date: | 2000-08-03 | | Release date: | 2000-08-23 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Mechanism of pH-dependent N-acetylgalactosamine binding by a functional mimic of the hepatocyte asialoglycoprotein receptor.
J.Biol.Chem., 275, 2000
|
|
2MLT
 
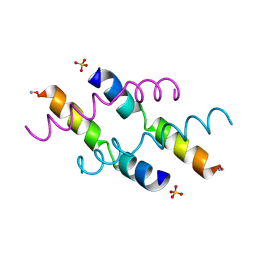 | |
3MB7
 
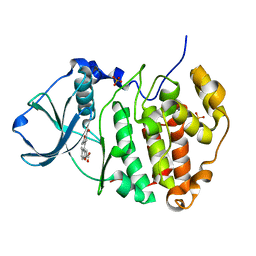 | | Human CK2 catalytic domain in complex with a difurane derivative inhibitor (AMR) | | Descriptor: | Casein kinase II subunit alpha, SULFATE ION, naphtho[2,1-b:7,8-b']difuran-2,9-dicarboxylic acid | | Authors: | Reiser, J.-B, Prudent, R, Cochet, C. | | Deposit date: | 2010-03-25 | | Release date: | 2010-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | New potent dual inhibitors of CK2 and Pim kinases: discovery and structural insights.
Faseb J., 24, 2010
|
|
4MOT
 
 | | Structure of Streptococcus pneumonia pare in complex with AZ13072886 | | Descriptor: | 1-[4-(3-methylbutyl)-5-oxo-6-(pyridin-3-yl)-4,5-dihydro[1,3]thiazolo[5,4-b]pyridin-2-yl]-3-prop-2-en-1-ylurea, Topoisomerase IV subunit B | | Authors: | Ogg, D, Boriack-Sjodin, P.A. | | Deposit date: | 2013-09-12 | | Release date: | 2013-11-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Thiazolopyridone ureas as DNA gyrase B inhibitors: Optimization of antitubercular activity and efficacy.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4MA9
 
 | |
4M49
 
 | | Lactate Dehydrogenase A in complex with a substituted pyrazine inhibitor compound 18 | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 3-(5-amino-6-{[(1R)-1-phenylethyl]amino}pyrazin-2-yl)-4-chlorobenzoic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Eigenbrot, C, Ultsch, M. | | Deposit date: | 2013-08-06 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | Identification of 2-amino-5-aryl-pyrazines as inhibitors of human lactate dehydrogenase.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
1YKR
 
 | | Crystal structure of cdk2 with an aminoimidazo pyridine inhibitor | | Descriptor: | 4-{[6-(2,6-DICHLOROBENZOYL)IMIDAZO[1,2-A]PYRIDIN-2-YL]AMINO}BENZENESULFONAMIDE, Cell division protein kinase 2 | | Authors: | Hamdouchi, C, Zhong, B, Mendoza, J, Jaramillo, C, Zhang, F, Brooks, H.B. | | Deposit date: | 2005-01-18 | | Release date: | 2006-01-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based design of a new class of highly selective aminoimidazo[1,2-a]pyridine-based inhibitors of cyclin dependent kinases
Bioorg.Med.Chem.Lett., 15, 2005
|
|
4JNK
 
 | | Lactate Dehydrogenase A in complex with inhibitor compound 22 | | Descriptor: | (2R)-2-{[5-cyano-4-(3,4-dichlorophenyl)-6-oxo-1,6-dihydropyrimidin-2-yl]sulfanyl}-N-(4-sulfamoylphenyl)propanamide, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Eigenbrot, C, Ultsch, M. | | Deposit date: | 2013-03-15 | | Release date: | 2013-05-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.896 Å) | | Cite: | Identification of substituted 2-thio-6-oxo-1,6-dihydropyrimidines as inhibitors of human lactate dehydrogenase.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4JPA
 
 | | Mmp13 in complex with a piperazine hydantoin ligand | | Descriptor: | 3-[({2-[4-({[(4S)-4-methyl-2,5-dioxoimidazolidin-4-yl]methyl}sulfonyl)piperazin-1-yl]pyrimidin-5-yl}oxy)methyl]benzonitrile, CALCIUM ION, Collagenase 3, ... | | Authors: | Gerhardt, S, Hargreaves, D. | | Deposit date: | 2013-03-19 | | Release date: | 2014-03-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Hydantoin based inhibitors of MMP13--discovery of AZD6605.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
1URC
 
 | | Cyclin A binding groove inhibitor Ace-Arg-Lys-Leu-Phe-Gly | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, CYCLIN A2, PEPTIDE INHIBITOR | | Authors: | Kontopidis, G, Andrews, M, McInnes, C, Cowan, A, Powers, H, Innes, L, Plater, A, Griffiths, G, Paterson, D, Zheleva, D, Lane, D, Green, S, Walkinshaw, M, Fischer, P. | | Deposit date: | 2003-10-28 | | Release date: | 2003-10-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Design, synthesis, biological activity and structural analysis of cyclic peptide inhibitors targeting the substrate recruitment site of cyclin-dependent kinase complexes.
Org. Biomol. Chem., 2, 2004
|
|
4JP4
 
 | | Mmp13 in complex with a reverse hydroxamate Zn-binder | | Descriptor: | CALCIUM ION, Collagenase 3, N-[(2S)-4-(5-fluoropyrimidin-2-yl)-1-({4-[5-(2,2,2-trifluoroethoxy)pyrimidin-2-yl]piperazin-1-yl}sulfonyl)butan-2-yl]-N-hydroxyformamide, ... | | Authors: | Gerhardt, S, Hargreaves, D. | | Deposit date: | 2013-03-19 | | Release date: | 2014-03-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Hydantoin based inhibitors of MMP13--discovery of AZD6605.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4N6U
 
 | | Adhiron: a stable and versatile peptide display scaffold - truncated adhiron | | Descriptor: | Adhiron | | Authors: | Mcpherson, M, Tomlinson, D, Owen, R.L, Nettleship, J.E, Owens, R.J. | | Deposit date: | 2013-10-14 | | Release date: | 2014-04-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Adhiron: a stable and versatile peptide display scaffold for molecular recognition applications.
Protein Eng.Des.Sel., 27, 2014
|
|
4N6T
 
 | | Adhiron: a stable and versatile peptide display scaffold - full length adhiron | | Descriptor: | Adhiron | | Authors: | Mcpherson, M, Tomlinson, D, Owen, R.L, Nettleship, J.E, Owens, R.J. | | Deposit date: | 2013-10-14 | | Release date: | 2014-04-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Adhiron: a stable and versatile peptide display scaffold for molecular recognition applications.
Protein Eng.Des.Sel., 27, 2014
|
|
3ICS
 
 | |
3ILQ
 
 | | Structure of mCD1d with bound glycolipid BbGL-2c from Borrelia burgdorferi | | Descriptor: | (2S)-3-(alpha-D-galactopyranosyloxy)-2-(hexadecanoyloxy)propyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zajonc, D.M. | | Deposit date: | 2009-08-07 | | Release date: | 2010-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Lipid binding orientation within CD1d affects recognition of Borrelia burgorferi antigens by NKT cells.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
