7NYT
 
 | | Trichoderma reesei Cel7A E212Q mutant in complex with lactose. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COBALT (II) ION, Exoglucanase 1, ... | | Authors: | Haataja, T, Sandgren, M, Stahlberg, J. | | Deposit date: | 2021-03-23 | | Release date: | 2022-03-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Enzyme kinetics by GH7 cellobiohydrolases on chromogenic substrates is dictated by non-productive binding: insights from crystal structures and MD simulation.
Febs J., 290, 2023
|
|
6USY
 
 | | COAGULATION FACTOR XI CATALYTIC DOMAIN (C123S) IN COMPLEX WITH NVP-XIV936 | | Descriptor: | 1-[(2S)-2-{3-[(3S)-3-amino-2,3-dihydro-1-benzofuran-5-yl]-5-(propan-2-yl)phenyl}-2-hydroxyethyl]-1H-indole-7-carboxylic acid, Coagulation factor XIa light chain | | Authors: | Weihofen, W.A, Clark, K, Nunes, S. | | Deposit date: | 2019-10-28 | | Release date: | 2020-07-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Structure-Based Design and Preclinical Characterization of Selective and Orally Bioavailable Factor XIa Inhibitors: Demonstrating the Power of an Integrated S1 Protease Family Approach.
J.Med.Chem., 63, 2020
|
|
7OFQ
 
 | |
7OC8
 
 | | Trichoderma reesei Cel7A E212Q mutant in complex with pNPL | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COBALT (II) ION, Exoglucanase 1, ... | | Authors: | Haataja, T, Sandgren, M, Stahlberg, J. | | Deposit date: | 2021-04-26 | | Release date: | 2022-03-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Enzyme kinetics by GH7 cellobiohydrolases on chromogenic substrates is dictated by non-productive binding: insights from crystal structures and MD simulation.
Febs J., 290, 2023
|
|
6VIZ
 
 | | BRD4_Bromodomain1 complex with pyrrolopyridone compound 27 | | Descriptor: | 4-[2-(2,6-dimethylphenoxy)-5-(ethylsulfonyl)phenyl]-N-ethyl-6-methyl-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridine-2-carboxamide, Bromodomain-containing protein 4 | | Authors: | Longenecker, K.L, Park, C.H, Qiu, W. | | Deposit date: | 2020-01-14 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.391 Å) | | Cite: | Discovery ofN-Ethyl-4-[2-(4-fluoro-2,6-dimethyl-phenoxy)-5-(1-hydroxy-1-methyl-ethyl)phenyl]-6-methyl-7-oxo-1H-pyrrolo[2,3-c]pyridine-2-carboxamide (ABBV-744), a BET Bromodomain Inhibitor with Selectivity for the Second Bromodomain.
J.Med.Chem., 63, 2020
|
|
6VIW
 
 | | BRD4_Bromodomain1 complex with pyrrolopyridone compound 18 | | Descriptor: | 4-[2-(2,4-difluorophenoxy)-5-(methylsulfonyl)phenyl]-N-ethyl-6-methyl-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridine-2-carboxamide, Bromodomain-containing protein 4 | | Authors: | Longenecker, K.L, Park, C.H, Qiu, W. | | Deposit date: | 2020-01-14 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.429 Å) | | Cite: | Discovery ofN-Ethyl-4-[2-(4-fluoro-2,6-dimethyl-phenoxy)-5-(1-hydroxy-1-methyl-ethyl)phenyl]-6-methyl-7-oxo-1H-pyrrolo[2,3-c]pyridine-2-carboxamide (ABBV-744), a BET Bromodomain Inhibitor with Selectivity for the Second Bromodomain.
J.Med.Chem., 63, 2020
|
|
6VIY
 
 | | BRD2_Bromodomain2 complex with pyrrolopyridone compound 27 | | Descriptor: | 4-[2-(2,6-dimethylphenoxy)-5-(ethylsulfonyl)phenyl]-N-ethyl-6-methyl-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridine-2-carboxamide, Bromodomain-containing protein 2 | | Authors: | Longenecker, K.L, Park, C.H, Qiu, W. | | Deposit date: | 2020-01-14 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.904 Å) | | Cite: | Discovery ofN-Ethyl-4-[2-(4-fluoro-2,6-dimethyl-phenoxy)-5-(1-hydroxy-1-methyl-ethyl)phenyl]-6-methyl-7-oxo-1H-pyrrolo[2,3-c]pyridine-2-carboxamide (ABBV-744), a BET Bromodomain Inhibitor with Selectivity for the Second Bromodomain.
J.Med.Chem., 63, 2020
|
|
6VIX
 
 | | BRD4_Bromodomain2 complex with pyrrolopyridone compound 18 | | Descriptor: | 4-[2-(2,4-difluorophenoxy)-5-(methylsulfonyl)phenyl]-N-ethyl-6-methyl-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridine-2-carboxamide, Bromodomain-containing protein 4 | | Authors: | Longenecker, K.L, Park, C.H, Qiu, W. | | Deposit date: | 2020-01-14 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.116 Å) | | Cite: | Discovery ofN-Ethyl-4-[2-(4-fluoro-2,6-dimethyl-phenoxy)-5-(1-hydroxy-1-methyl-ethyl)phenyl]-6-methyl-7-oxo-1H-pyrrolo[2,3-c]pyridine-2-carboxamide (ABBV-744), a BET Bromodomain Inhibitor with Selectivity for the Second Bromodomain.
J.Med.Chem., 63, 2020
|
|
9FDB
 
 | | Co-crystal structure of Galectin-3 with an inhibitor | | Descriptor: | (2~{R},3~{R},4~{S},5~{R},6~{R})-~{N}-cyclopropyl-~{N}-(2-hydroxyethyl)-6-(hydroxymethyl)-3,5-bis(oxidanyl)-4-[4-[3,4,5-tris(fluoranyl)phenyl]-1,2,3-triazol-1-yl]oxane-2-carboxamide, Galectin-3, THIOCYANATE ION | | Authors: | Mac Sweeney, A. | | Deposit date: | 2024-05-16 | | Release date: | 2025-03-26 | | Last modified: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Discovery of Galactopyranose-1-carboxamides as a New Class of Small, Novel, Potent, Selective, and Orally Active Galectin-3 Inhibitors.
Chemmedchem, 20, 2025
|
|
9FDC
 
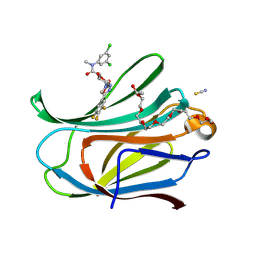 | | Co-crystal structure of Galectin-3 with an inhibitor | | Descriptor: | (2~{R},3~{R},4~{S},5~{R},6~{R})-~{N}-[3,5-bis(chloranyl)phenyl]-6-(hydroxymethyl)-~{N}-methyl-3,5-bis(oxidanyl)-4-[4-[3,4,5-tris(fluoranyl)phenyl]-1,2,3-triazol-1-yl]oxane-2-carboxamide, Galectin-3, NONAETHYLENE GLYCOL, ... | | Authors: | Mac Sweeney, A. | | Deposit date: | 2024-05-16 | | Release date: | 2025-03-26 | | Last modified: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Discovery of Galactopyranose-1-carboxamides as a New Class of Small, Novel, Potent, Selective, and Orally Active Galectin-3 Inhibitors.
Chemmedchem, 20, 2025
|
|
6QMR
 
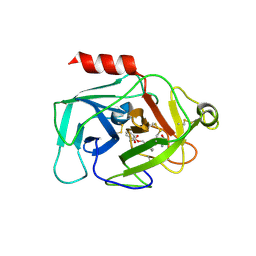 | | Complement factor D in complex with the inhibitor (S)-2-(2-((3'-(1-amino-2-hydroxyethyl)-[1,1'-biphenyl]-3-yl)methoxy)phenyl)acetic acid | | Descriptor: | 2-[2-[[3-[3-[(1~{S})-1-azanyl-2-oxidanyl-ethyl]phenyl]phenyl]methoxy]phenyl]ethanoic acid, Complement factor D | | Authors: | Karki, R, Powers, J, Mainolfi, N, Anderson, K, Belanger, D, Liu, D, Jendza, K, Gelin, C.F, Solovay, C, Mac Sweeeny, A, Delgado, O, Crowley, M, Liao, S.-M, Argikar, U.A, Flohr, S, La Bonte, L.R, Lorthiois, E.L, Vulpetti, A, Cumin, F, Brown, A, Adams, C, Jaffee, B, Mogi, M. | | Deposit date: | 2019-02-08 | | Release date: | 2019-04-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design, Synthesis, and Preclinical Characterization of Selective Factor D Inhibitors Targeting the Alternative Complement Pathway.
J.Med.Chem., 62, 2019
|
|
7VC6
 
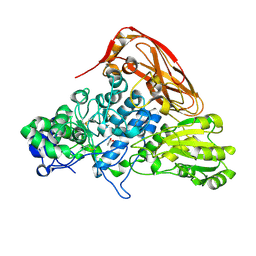 | | The structure of beta-xylosidase from Phanerochaete chrysosporium(PcBxl3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, xylan 1,4-beta-xylosidase | | Authors: | Kojima, K, Sunagawa, N, Igarashi, K. | | Deposit date: | 2021-09-01 | | Release date: | 2022-02-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Comparison of glycoside hydrolase family 3 beta-xylosidases from basidiomycetes and ascomycetes reveals evolutionarily distinct xylan degradation systems.
J.Biol.Chem., 298, 2022
|
|
7VC7
 
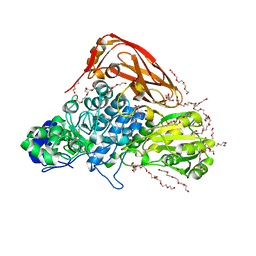 | | The structure of beta-xylosidase from Phanerochaete chrysosporium(PcBxl3) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kojima, K, Sunagawa, N, Igarashi, K. | | Deposit date: | 2021-09-01 | | Release date: | 2022-02-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Comparison of glycoside hydrolase family 3 beta-xylosidases from basidiomycetes and ascomycetes reveals evolutionarily distinct xylan degradation systems.
J.Biol.Chem., 298, 2022
|
|
4X62
 
 | | Crystal Structure of 30S ribosomal subunit from Thermus thermophilus | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Demirci, H, Chen, J, Choi, J, Soltis, M, Puglisi, J.D. | | Deposit date: | 2014-12-06 | | Release date: | 2015-11-18 | | Last modified: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (3.4492 Å) | | Cite: | N(6)-methyladenosine in mRNA disrupts tRNA selection and translation-elongation dynamics.
Nat.Struct.Mol.Biol., 23, 2016
|
|
4X65
 
 | | Crystal Structure of 30S ribosomal subunit from Thermus thermophilus | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Demirci, H, Chen, J, Choi, J, Soltis, M, Puglisi, J.D. | | Deposit date: | 2014-12-06 | | Release date: | 2015-11-18 | | Last modified: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (3.345 Å) | | Cite: | N(6)-methyladenosine in mRNA disrupts tRNA selection and translation-elongation dynamics.
Nat.Struct.Mol.Biol., 23, 2016
|
|
8P8M
 
 | | Yeast 60S ribosomal subunit, RPL39 deletion | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Rabl, J, Banerjee, A, Boehringer, D, Zavolan, M. | | Deposit date: | 2023-06-01 | | Release date: | 2024-06-12 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Ribosomal protein RPL39L is an efficiency factor in the cotranslational folding of a subset of proteins with alpha helical domains.
Nucleic Acids Res., 52, 2024
|
|
8PFR
 
 | | Mouse RPL39L integrated into the yeast 60S ribosomal subunit | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Rabl, J, Banerjee, A, Boehringer, D, Zavolan, M. | | Deposit date: | 2023-06-16 | | Release date: | 2024-06-26 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.15 Å) | | Cite: | Ribosomal protein RPL39L is an efficiency factor in the cotranslational folding of a subset of proteins with alpha helical domains.
Nucleic Acids Res., 52, 2024
|
|
8P8U
 
 | | Yeast 60S ribosomal subunit | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Rabl, J, Banerjee, A, Boehringer, D, Zavolan, M. | | Deposit date: | 2023-06-02 | | Release date: | 2024-06-12 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.23 Å) | | Cite: | Ribosomal protein RPL39L is an efficiency factor in the cotranslational folding of a subset of proteins with alpha helical domains.
Nucleic Acids Res., 52, 2024
|
|
8P8N
 
 | | Mouse RPL39 integrated into the yeast 60S ribosomal subunit | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Rabl, J, Banerjee, A, Boehringer, D, Zavolan, M. | | Deposit date: | 2023-06-02 | | Release date: | 2024-06-12 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.15 Å) | | Cite: | Ribosomal protein RPL39L is an efficiency factor in the cotranslational folding of a subset of proteins with alpha helical domains.
Nucleic Acids Res., 52, 2024
|
|
6R89
 
 | | Structure of Arabidopsis thaliana GLR3.3 ligand-binding domain in complex with L-cysteine | | Descriptor: | CHLORIDE ION, CYSTEINE, GLYCEROL, ... | | Authors: | Alfieri, A, Pederzoli, R, Costa, A. | | Deposit date: | 2019-04-01 | | Release date: | 2020-01-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structural bases for agonist diversity in anArabidopsis thalianaglutamate receptor-like channel.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3EJ1
 
 | | CDK2/CyclinA complexed with a pyrazolopyridazine inhibitor | | Descriptor: | Cell division protein kinase 2, Cyclin-A2, N-cyclopropyl-4-pyrazolo[1,5-b]pyridazin-3-ylpyrimidin-2-amine | | Authors: | Stevens, K, Reno, M, Alberti, J, Price, D, Kane-Carson, L, Knick, V, Shewchuk, L, Hassell, A, Veal, J, Peel, M. | | Deposit date: | 2008-09-17 | | Release date: | 2008-10-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Synthesis and evaluation of pyrazolo[1,5-b]pyridazines as selective cyclin dependent kinase inhibitors.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
4NMG
 
 | |
4NLF
 
 | |
6GEV
 
 | | Mineralocorticoid receptor in complex with (s)-13 | | Descriptor: | 6-[[(3~{S})-7-fluoranyl-3-(2-methylpropyl)-2,3-dihydro-1,4-benzoxazin-4-yl]carbonyl]-4~{H}-1,4-benzoxazin-3-one, GLYCEROL, Mineralocorticoid receptor, ... | | Authors: | Edman, K, Aagaard, A, Tangefjord, S. | | Deposit date: | 2018-04-27 | | Release date: | 2019-01-09 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Identification of Mineralocorticoid Receptor Modulators with Low Impact on Electrolyte Homeostasis but Maintained Organ Protection.
J.Med.Chem., 62, 2019
|
|
3EID
 
 | | CDK2/CyclinA complexed with a pyrazolopyridazine inhibitor | | Descriptor: | (2S)-1-(dimethylamino)-3-(4-{[4-(6-morpholin-4-ylpyrazolo[1,5-b]pyridazin-3-yl)pyrimidin-2-yl]amino}phenoxy)propan-2-ol, Cell division protein kinase 2, Cyclin-A2 | | Authors: | Steven, K, Reno, M, Alberti, J, Price, D, Kane-Carson, L, Knick, V, Shewchuk, L, Hassell, A, Veal, J, Peel, M. | | Deposit date: | 2008-09-15 | | Release date: | 2008-10-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Synthesis and evaluation of pyrazolo[1,5-b]pyridazines as selective cyclin dependent kinase inhibitors.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
