6OMN
 
 | | Glycosylated BMP2 homodimer | | Descriptor: | Bone morphogenetic protein 2, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Juo, Z.S, Seeherman, H. | | Deposit date: | 2019-04-19 | | Release date: | 2019-05-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | A BMP/activin A chimera is superior to native BMPs and induces bone repair in nonhuman primates when delivered in a composite matrix.
Sci Transl Med, 11, 2019
|
|
6OML
 
 | | Human BMP chimera BV261 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Bone morphogenetic protein 2 and Bone morphogenetic protein 6, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Seeherman, H, Juo, Z.S. | | Deposit date: | 2019-04-18 | | Release date: | 2019-05-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A BMP/activin A chimera is superior to native BMPs and induces bone repair in nonhuman primates when delivered in a composite matrix.
Sci Transl Med, 11, 2019
|
|
2R82
 
 | | Pyruvate phosphate dikinase (PPDK) triple mutant R219E/E271R/S262D adapts a second conformational state | | Descriptor: | Pyruvate, phosphate dikinase, SULFATE ION | | Authors: | Lim, K, Read, R.J, Chen, C.C, Herzberg, O. | | Deposit date: | 2007-09-10 | | Release date: | 2008-01-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Swiveling domain mechanism in pyruvate phosphate dikinase.
Biochemistry, 46, 2007
|
|
2M7T
 
 | | Solution NMR Structure of Engineered Cystine Knot Protein 2.5D | | Descriptor: | Cystine Knot Protein 2.5D | | Authors: | Cochran, F.V, Das, R. | | Deposit date: | 2013-04-30 | | Release date: | 2014-05-07 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Challenging the state of the art in protein structure prediction: Highlights of experimental target structures for the 10th Critical Assessment of Techniques for Protein Structure Prediction Experiment CASP10.
Proteins, 82 Suppl 2, 2014
|
|
6W9H
 
 | | SUBSTITUTED BENZYLOXYTRICYCLIC COMPOUNDS AS RETINOIC ACID-RELATED ORPHAN RECEPTOR GAMMA T AGONISTS | | Descriptor: | 4-{(3R)-3-[4-(benzyloxy)phenyl]-3-[(4-fluorophenyl)sulfonyl]pyrrolidine-1-carbonyl}-1lambda~6~-thiane-1,1-dione, Nuclear receptor ROR-gamma, Steroid receptor coactivator 1 fusion | | Authors: | Sack, J.S. | | Deposit date: | 2020-03-23 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.995 Å) | | Cite: | Substituted benzyloxytricyclic compounds as retinoic acid-related orphan receptor gamma t (ROR gamma t) agonists.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
6W9I
 
 | |
5WJ8
 
 | | Crystal Structure of Human Cadherin-23 EC13-14 | | Descriptor: | CALCIUM ION, CHLORIDE ION, Cadherin-23, ... | | Authors: | Velez-Cortes, F, Conghui, C, De-la-Torre, P, Sotomayor, M. | | Deposit date: | 2017-07-21 | | Release date: | 2018-07-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Zooming in on Cadherin-23: Structural Diversity and Potential Mechanisms of Inherited Deafness.
Structure, 26, 2018
|
|
3TWO
 
 | | The crystal structure of CAD from Helicobacter pylori complexed with NADP(H) | | Descriptor: | Mannitol dehydrogenase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ZINC ION | | Authors: | Seo, K.H, Zhuang, N.N, Cong, C, Lee, K.H. | | Deposit date: | 2011-09-22 | | Release date: | 2011-10-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Unusual NADPH conformation in the crystal structure of a cinnamyl alcohol dehydrogenase from Helicobacter pylori in complex with NADP(H) and substrate docking analysis
Febs Lett., 586, 2012
|
|
4AE1
 
 | |
4AE0
 
 | |
2O14
 
 | | X-Ray Crystal Structure of Protein YXIM_BACsu from Bacillus subtilis. Northeast Structural Genomics Consortium Target SR595 | | Descriptor: | Hypothetical protein yxiM, MANGANESE (II) ION, SULFATE ION | | Authors: | Kuzin, A.P, Chen, Y, Seetharaman, J, Chen, C.X, Fang, Y, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-11-28 | | Release date: | 2006-12-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-Ray structure of the hypothetical protein YXIM_BACsu from Bacillus subtilis.
To be Published
|
|
7CYC
 
 | | Cryo-EM structures of Alphacoronavirus spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Song, X, Shi, Y, Ding, W, Liu, Z.J, Peng, G. | | Deposit date: | 2020-09-03 | | Release date: | 2020-12-23 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Cryo-EM analysis of the HCoV-229E spike glycoprotein reveals dynamic prefusion conformational changes.
Nat Commun, 12, 2021
|
|
4I1L
 
 | | Structural and Biological Features of FOXP3 Dimerization Relevant to Regulatory T Cell Function | | Descriptor: | ACETATE ION, Forkhead box protein P3, MAGNESIUM ION, ... | | Authors: | Song, X.M, Greene, M.I, Zhou, Z.C. | | Deposit date: | 2012-11-21 | | Release date: | 2012-12-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and biological features of FOXP3 dimerization relevant to regulatory T cell function.
Cell Rep, 1, 2012
|
|
7CYD
 
 | | Cryo-EM structures of Alphacoronavirus spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Song, X, Shi, Y, Ding, W, Liu, Z.J, Peng, G. | | Deposit date: | 2020-09-03 | | Release date: | 2020-12-23 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Cryo-EM analysis of the HCoV-229E spike glycoprotein reveals dynamic prefusion conformational changes.
Nat Commun, 12, 2021
|
|
4LGI
 
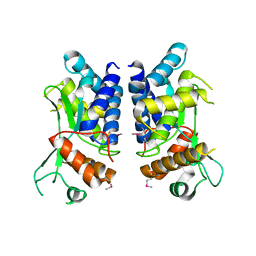 | | N-terminal truncated NleC structure | | Descriptor: | Uncharacterized protein | | Authors: | Li, W.Q, Liu, Y.X, Sheng, X.L, Yan, C.Y, Wang, J.W. | | Deposit date: | 2013-06-28 | | Release date: | 2014-01-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structure and mechanism of a type III secretion protease, NleC
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7KMK
 
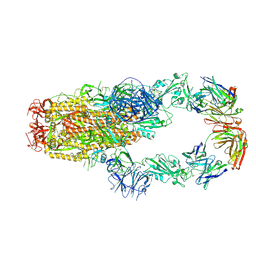 | | cryo-EM structure of SARS-CoV-2 spike in complex with Fab 15033-7, two RBDs bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 15033-7 heavy chain, ... | | Authors: | Li, Z, Rini, J.M. | | Deposit date: | 2020-11-03 | | Release date: | 2021-02-10 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Tetravalent SARS-CoV-2 Neutralizing Antibodies Show Enhanced Potency and Resistance to Escape Mutations.
J.Mol.Biol., 433, 2021
|
|
7KLG
 
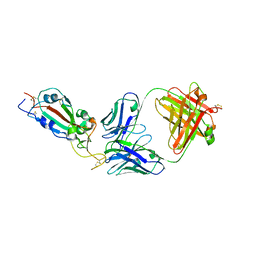 | | SARS-CoV-2 RBD in complex with Fab 15033 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 15033 heavy chain, Fab 15033 light chain, ... | | Authors: | Li, Z, Rini, J.M. | | Deposit date: | 2020-10-30 | | Release date: | 2021-02-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Tetravalent SARS-CoV-2 Neutralizing Antibodies Show Enhanced Potency and Resistance to Escape Mutations.
J.Mol.Biol., 433, 2021
|
|
7KML
 
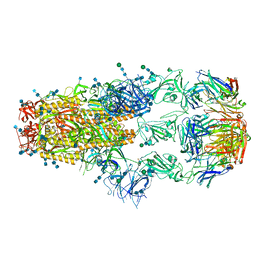 | | cryo-EM structure of SARS-CoV-2 spike in complex with Fab 15033-7, three RBDs bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 15033-7 heavy chain, ... | | Authors: | Li, Z, Rini, J.M. | | Deposit date: | 2020-11-03 | | Release date: | 2021-02-10 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Tetravalent SARS-CoV-2 Neutralizing Antibodies Show Enhanced Potency and Resistance to Escape Mutations.
J.Mol.Biol., 433, 2021
|
|
7KLH
 
 | | SARS-CoV-2 RBD in complex with Fab 15033-7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 15033-7 heavy chain, Fab 15033-7 light chain, ... | | Authors: | Li, Z, Rini, J.M. | | Deposit date: | 2020-10-30 | | Release date: | 2021-02-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Tetravalent SARS-CoV-2 Neutralizing Antibodies Show Enhanced Potency and Resistance to Escape Mutations.
J.Mol.Biol., 433, 2021
|
|
8EQN
 
 | |
7JKS
 
 | | Crystal structure of vaccine-elicited broadly neutralizing VRC01-class antibody 2411a in complex with HIV-1 gp120 core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 gp120 core, The heavy chain of antibody 2411a, ... | | Authors: | Zhou, T, Chen, X, Kwong, P.D, Mascola, J.R. | | Deposit date: | 2020-07-28 | | Release date: | 2021-06-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Vaccination induces maturation in a mouse model of diverse unmutated VRC01-class precursors to HIV-neutralizing antibodies with >50% breadth.
Immunity, 54, 2021
|
|
7JKT
 
 | | Crystal structure of vaccine-elicited broadly neutralizing VRC01-class antibody 2413a in complex with HIV-1 gp120 core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HIV-1 gp120 core from strain d45-01dG5, ... | | Authors: | Zhou, T, Chen, X, Kwong, P.D, Mascola, J.R. | | Deposit date: | 2020-07-28 | | Release date: | 2021-06-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Vaccination induces maturation in a mouse model of diverse unmutated VRC01-class precursors to HIV-neutralizing antibodies with >50% breadth.
Immunity, 54, 2021
|
|
8HJT
 
 | | Crystal Structure of Intracellular B30.2 Domain of VpBTN3 and VpBTN2 in Complex with HMBPP | | Descriptor: | (2E)-4-hydroxy-3-methylbut-2-en-1-yl trihydrogen diphosphate, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Butyrophylin 3, ... | | Authors: | Yang, Y.Y, Shen, P.P, Li, X, Yi, S.M, Zhang, M.T, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-11-23 | | Release date: | 2023-09-13 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Phosphoantigens glue butyrophilin 3A1 and 2A1 to activate V gamma 9V delta 2 T cells.
Nature, 621, 2023
|
|
5F7E
 
 | | Crystal structure of germ-line precursor of 3BNC60 Fab | | Descriptor: | Fab heavy chain, Fab light chain | | Authors: | Sievers, S.A, Scharf, L, Jiang, S, Bjorkman, P.J. | | Deposit date: | 2015-12-08 | | Release date: | 2016-04-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for germline antibody recognition of HIV-1 immunogens.
Elife, 5, 2016
|
|
5B6D
 
 | |
