5FMQ
 
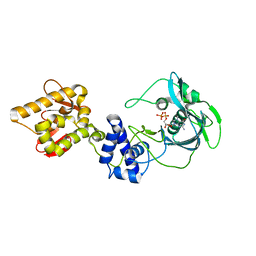 | | Crystal structure of the mid, cap-binding, mid-link and 627 domains from avian influenza A virus polymerase PB2 subunit bound to M7GTP H32 crystal form | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, INFLUENZA A PB2 SUBUNIT | | Authors: | Thierry, E, Pflug, A, Hart, D, Cusack, S. | | Deposit date: | 2015-11-07 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Influenza Polymerase Can Adopt an Alternative Configuration Involving a Radical Repacking of Pb2 Domains.
Mol.Cell, 61, 2016
|
|
5FML
 
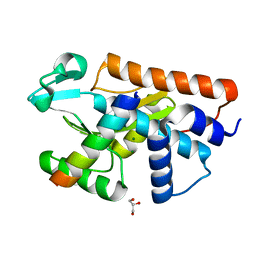 | | Crystal structure of the endonuclease from the PA subunit of influenza B virus bound to the PB2 subunit NLS peptide | | Descriptor: | GLYCEROL, MAGNESIUM ION, PA SUBUNIT OF INFLUENZA B POLYMERASE, ... | | Authors: | Guilligay, D, Gaudon, S, Cusack, S. | | Deposit date: | 2015-11-06 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Influenza Polymerase Can Adopt an Alternative Configuration Involving a Radical Repacking of Pb2 Domains.
Mol.Cell, 61, 2016
|
|
5FM6
 
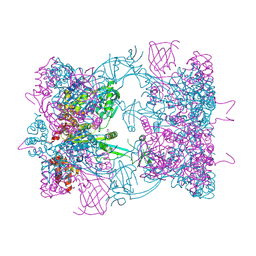 | | Double-heterohexameric rings of full-length Rvb1(ADP)Rvb2(apo) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, PHOSPHATE ION, RVB1, ... | | Authors: | Silva-Martin, N, Dauden, M.I, Glatt, S, Hoffmann, N.A, Mueller, C.W. | | Deposit date: | 2015-11-02 | | Release date: | 2016-01-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.997 Å) | | Cite: | The Combination of X-Ray Crystallography and Cryo-Electron Microscopy Provides Insight Into the Overall Architecture of the Dodecameric Rvb1/Rvb2 Complex.
Plos One, 11, 2016
|
|
5VNW
 
 | |
5T4X
 
 | | CRYSTAL STRUCTURE OF PDE6D IN APO-STATE | | Descriptor: | Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit delta | | Authors: | Qureshi, B.M, Schmidt, A, Scheerer, P. | | Deposit date: | 2016-08-30 | | Release date: | 2018-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Mechanistic insights into the role of prenyl-binding protein PrBP/ delta in membrane dissociation of phosphodiesterase 6.
Nat Commun, 9, 2018
|
|
1G9R
 
 | | CRYSTAL STRUCTURE OF GALACTOSYLTRANSFERASE LGTC IN COMPLEX WITH MN AND UDP-2F-GALACTOSE | | Descriptor: | ACETIC ACID, GLYCOSYL TRANSFERASE, MANGANESE (II) ION, ... | | Authors: | Persson, K, Hoa, D.L, Diekelmann, M, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C.J. | | Deposit date: | 2000-11-27 | | Release date: | 2001-02-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the retaining galactosyltransferase LgtC from Neisseria meningitidis in complex with donor and acceptor sugar analogs.
Nat.Struct.Biol., 8, 2001
|
|
1GA8
 
 | | CRYSTAL STRUCTURE OF GALACOSYLTRANSFERASE LGTC IN COMPLEX WITH DONOR AND ACCEPTOR SUGAR ANALOGS. | | Descriptor: | 4-deoxy-beta-D-xylo-hexopyranose-(1-4)-beta-D-glucopyranose, GALACTOSYL TRANSFERASE LGTC, MANGANESE (II) ION, ... | | Authors: | Persson, K, Ly, H.D, Diekelmann, M, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C.J. | | Deposit date: | 2000-11-29 | | Release date: | 2001-02-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the retaining galactosyltransferase LgtC from Neisseria meningitidis in complex with donor and acceptor sugar analogs.
Nat.Struct.Biol., 8, 2001
|
|
9HLK
 
 | | X-ray structure of the adduct formed upon reaction of the diiodido analogue of picoplatin with lysozyme (structure A) | | Descriptor: | ACETATE ION, AMMONIA, CHLORIDE ION, ... | | Authors: | Ferraro, G, Merlino, A. | | Deposit date: | 2024-12-05 | | Release date: | 2025-05-14 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Cytotoxicity and Binding to DNA, Lysozyme, Ribonuclease A, and Human Serum Albumin of the Diiodido Analog of Picoplatin.
Inorg.Chem., 64, 2025
|
|
9HN6
 
 | | X-ray structure of the adduct formed upon reaction of the diiodido analogue of picoplatin with ribonuclease A | | Descriptor: | AMMONIA, CHLORIDE ION, IODIDE ION, ... | | Authors: | Ferraro, G, Merlino, A. | | Deposit date: | 2024-12-10 | | Release date: | 2025-05-14 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Cytotoxicity and Binding to DNA, Lysozyme, Ribonuclease A, and Human Serum Albumin of the Diiodido Analog of Picoplatin.
Inorg.Chem., 64, 2025
|
|
9HNB
 
 | |
9HMK
 
 | | X-ray structure of the adduct formed upon reaction of the diiodido analogue of picoplatin with lysozyme (structure B) | | Descriptor: | IODIDE ION, Lysozyme C, PLATINUM (II) ION | | Authors: | Ferraro, G, Merlino, A. | | Deposit date: | 2024-12-09 | | Release date: | 2025-05-14 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Cytotoxicity and Binding to DNA, Lysozyme, Ribonuclease A, and Human Serum Albumin of the Diiodido Analog of Picoplatin.
Inorg.Chem., 64, 2025
|
|
9HMQ
 
 | | X-structure of the adduct formed upon reaction of the diiodido analogue of picoplatin with lysozyme (structure C) | | Descriptor: | AMMONIA, IODIDE ION, Lysozyme C, ... | | Authors: | Ferraro, G, Merlino, A. | | Deposit date: | 2024-12-09 | | Release date: | 2025-05-14 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Cytotoxicity and Binding to DNA, Lysozyme, Ribonuclease A, and Human Serum Albumin of the Diiodido Analog of Picoplatin.
Inorg.Chem., 64, 2025
|
|
2HOR
 
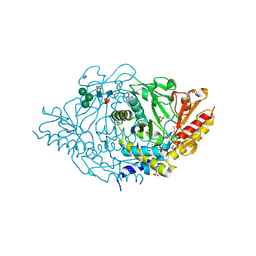 | | Crystal structure of alliinase from garlic- apo form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Alliin lyase 1, ... | | Authors: | Shimon, L.J.W, Rabinkov, A, Wilcheck, M, Mirelman, D, Frolow, F. | | Deposit date: | 2006-07-16 | | Release date: | 2007-02-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Two Structures of Alliinase from Alliium sativum L.: Apo Form and Ternary Complex with Aminoacrylate Reaction Intermediate Covalently Bound to the PLP Cofactor.
J.Mol.Biol., 366, 2007
|
|
4FMT
 
 | |
9H1F
 
 | |
5LAX
 
 | | Crystal structure of HLA_DRB1*04:01 in complex with alpha-enolase peptide 26-40 | | Descriptor: | HLA class II histocompatibility antigen, DR alpha chain, DRB1-4 beta chain, ... | | Authors: | Dubnovitsky, A, Kozhukh, G, Sandalova, T, Achour, A. | | Deposit date: | 2016-06-15 | | Release date: | 2016-12-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Functional and Structural Characterization of a Novel HLA-DRB1*04:01-Restricted alpha-Enolase T Cell Epitope in Rheumatoid Arthritis.
Front Immunol, 7, 2016
|
|
8Q2M
 
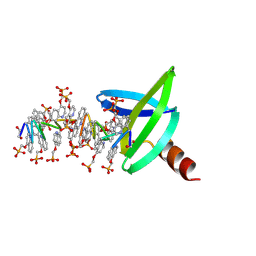 | | 18mer DNA mimic Foldamer with an Aliphatic linker in complex with Sac7d V26A/M29A protein | | Descriptor: | DNA mimic Foldamer, DNA-binding protein 7b | | Authors: | Deepak, D, Corvaglia, V, Wu, J, Huc, I. | | Deposit date: | 2023-08-02 | | Release date: | 2023-08-23 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | DNA Mimic Foldamer Recognition of a Chromosomal Protein.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
5M2N
 
 | |
4PY7
 
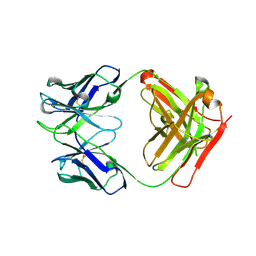 | | Crystal Structure of Fab 3.1 | | Descriptor: | antibody 3.1 heavy chain, antibody 3.1 light chain | | Authors: | Dreyfus, C. | | Deposit date: | 2014-03-26 | | Release date: | 2014-05-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Alternative Recognition of the Conserved Stem Epitope in Influenza A Virus Hemagglutinin by a VH3-30-Encoded Heterosubtypic Antibody.
J.Virol., 88, 2014
|
|
4PY8
 
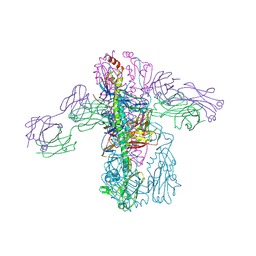 | | Crystal structure of Fab 3.1 in complex with the 1918 influenza virus hemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, ... | | Authors: | Dreyfus, C. | | Deposit date: | 2014-03-26 | | Release date: | 2014-05-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Alternative Recognition of the Conserved Stem Epitope in Influenza A Virus Hemagglutinin by a VH3-30-Encoded Heterosubtypic Antibody.
J.Virol., 88, 2014
|
|
6QD6
 
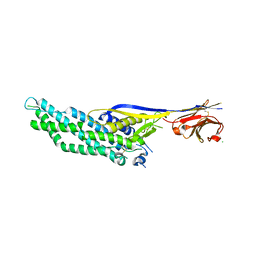 | | Molecular scaffolds expand the nanobody toolkit for cryo-EM applications: crystal structure of Mb-cHopQ-Nb207 | | Descriptor: | CHLORIDE ION, Mb-cHopQ-Nb207,Outer membrane protein,Mb-cHopQ-Nb207,Outer membrane protein,Mb-cHopQ-Nb207 | | Authors: | Uchanski, T, Masiulis, S, Fischer, B, Kalichuk, V, Wohlkonig, A, Zogg, T, Remaut, H, Vranken, W, Aricescu, A.R, Pardon, E, Steyaert, J. | | Deposit date: | 2018-12-31 | | Release date: | 2019-12-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Megabodies expand the nanobody toolkit for protein structure determination by single-particle cryo-EM
Nat.Methods, 18, 2021
|
|
6QFA
 
 | | CryoEM structure of a beta3K279T GABA(A)R homomer in complex with histamine and megabody Mb25 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid receptor subunit beta-3,Gamma-aminobutyric acid receptor subunit beta-3, HISTAMINE, ... | | Authors: | Uchanski, T, Masiulis, S, Fischer, B, Kalichuk, V, Wohlkoening, A, Zoegg, T, Remaut, H, Vranken, W, Aricescu, A.R, Pardon, E, Steyaert, J. | | Deposit date: | 2019-01-09 | | Release date: | 2021-08-04 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Megabodies expand the nanobody toolkit for protein structure determination by single-particle cryo-EM.
Nat.Methods, 18, 2021
|
|
5AFF
 
 | | Symportin 1 chaperones 5S RNP assembly during ribosome biogenesis by occupying an essential rRNA binding site | | Descriptor: | RIBOSOMAL PROTEIN L11, RIBOSOMAL PROTEIN L5, SYMPORTIN 1 | | Authors: | Calvino, F.R, Kharde, S, Wild, K, Bange, G, Sinning, I. | | Deposit date: | 2015-01-21 | | Release date: | 2015-04-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.398 Å) | | Cite: | Symportin 1 Chaperones 5S Rnp Assembly During Ribosome Biogenesis by Occupying an Essential Rrna-Binding Site.
Nat.Commun., 6, 2015
|
|
6RM9
 
 | | Crystal structure of the DEAH-box ATPase Prp2 in complex with Spp2 and ADP | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Hamann, F, Neumann, P, Ficner, R. | | Deposit date: | 2019-05-06 | | Release date: | 2020-02-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural analysis of the intrinsically disordered splicing factor Spp2 and its binding to the DEAH-box ATPase Prp2.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4RIE
 
 | | Landomycin Glycosyltransferase LanGT2 | | Descriptor: | Glycosyl transferase homolog | | Authors: | Tam, H.K, Gerhardt, S, Breit, B, Bechthold, A, Einsle, O. | | Deposit date: | 2014-10-06 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.162 Å) | | Cite: | Structural Characterization of O- and C-Glycosylating Variants of the Landomycin Glycosyltransferase LanGT2.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
