6FFZ
 
 | | Crystal structure of R. ruber ADH-A, mutant F43H, Y54L | | Descriptor: | Alcohol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | Authors: | Dobritzsch, D, Maurer, D, Hamnevik, E, Enugala, T.R, Widersten, M. | | Deposit date: | 2018-01-09 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Directed Evolution of Alcohol Dehydrogenase for Improved Stereoselective Redox Transformations of 1-Phenylethane-1,2-diol and Its Corresponding Acyloin.
Biochemistry, 57, 2018
|
|
6FFX
 
 | | Crystal structure of R. ruber ADH-A, mutant F43H | | Descriptor: | Alcohol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | Authors: | Dobritzsch, D, Maurer, D, Hamnevik, E, Enugala, T.R, Widersten, M. | | Deposit date: | 2018-01-09 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Directed Evolution of Alcohol Dehydrogenase for Improved Stereoselective Redox Transformations of 1-Phenylethane-1,2-diol and Its Corresponding Acyloin.
Biochemistry, 57, 2018
|
|
7QEI
 
 | | Structure of human MTHFD2L in complex with TH7299 | | Descriptor: | (2S)-2-[[4-[[2,4-bis(azanyl)-6-oxidanylidene-1H-pyrimidin-5-yl]carbamoylamino]phenyl]carbonylamino]pentanedioic acid, 2'-MONOPHOSPHOADENOSINE-5'-DIPHOSPHATE, Probable bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase 2 | | Authors: | Gustafsson, R, Scaletti, E.R, Stenmark, P. | | Deposit date: | 2021-12-03 | | Release date: | 2022-10-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The First Structure of Human MTHFD2L and Its Implications for the Development of Isoform-Selective Inhibitors.
Chemmedchem, 17, 2022
|
|
7PNM
 
 | |
7PO5
 
 | |
7PNQ
 
 | |
7DV4
 
 | | Crystal structure of anti-CTLA-4 VH domain in complex with human CTLA-4 | | Descriptor: | 1,2-ETHANEDIOL, 4003-1(VH), Cytotoxic T-lymphocyte protein 4 | | Authors: | Li, H, Gan, X, He, Y. | | Deposit date: | 2021-01-12 | | Release date: | 2022-01-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | An anti-CTLA-4 heavy chain-only antibody with enhanced T reg depletion shows excellent preclinical efficacy and safety profile.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
4B3H
 
 | |
4A2Z
 
 | | CRYSTAL STRUCTURE OF LEISHMANIA MAJOR N-MYRISTOYLTRANSFERASE (NMT) WITH BOUND MYRISTOYL-COA AND A PYRAZOLE SULPHONAMIDE LIGAND | | Descriptor: | 4-METHOXY-2,3,6-TRIMETHYL-N-(1,3,5-TRIMETHYL-1H-PYRAZOL-4-YL)BENZENESULFONAMIDE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Robinson, D.A, Brand, S, Fairlamb, A.H, Ferguson, M.A.J, Frearson, J.A, Wyatt, P.G. | | Deposit date: | 2011-09-29 | | Release date: | 2011-12-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Discovery of a novel class of orally active trypanocidal N-myristoyltransferase inhibitors.
J. Med. Chem., 55, 2012
|
|
8B5S
 
 | | Crystal Structure of P. aeruginosa WaaG in complex with UDP-glucose | | Descriptor: | UDP-glucose:(Heptosyl) LPS alpha 1,3-glucosyltransferase WaaG, URIDINE-5'-DIPHOSPHATE, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Scaletti, E, Gustafsson Westergren, R, Stenmark, P. | | Deposit date: | 2022-09-24 | | Release date: | 2023-10-04 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and functional insights into the Pseudomonas aeruginosa glycosyltransferase WaaG and the implications for lipopolysaccharide biosynthesis.
J.Biol.Chem., 299, 2023
|
|
8B5Q
 
 | |
8B62
 
 | | Crystal Structure of P. aeruginosa WaaG in complex with UDP-galactose | | Descriptor: | GALACTOSE-URIDINE-5'-DIPHOSPHATE, GLYCEROL, UDP-glucose:(Heptosyl) LPS alpha 1,3-glucosyltransferase WaaG | | Authors: | Scaletti, E, Gustafsson Westergren, R, Stenmark, P. | | Deposit date: | 2022-09-25 | | Release date: | 2023-10-04 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural and functional insights into the Pseudomonas aeruginosa glycosyltransferase WaaG and the implications for lipopolysaccharide biosynthesis.
J.Biol.Chem., 299, 2023
|
|
8B63
 
 | | Crystal Structure of P. aeruginosa WaaG in complex with UDP-GalNAc | | Descriptor: | ACETATE ION, UDP-glucose:(Heptosyl) LPS alpha 1,3-glucosyltransferase WaaG, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Scaletti, E, Gustafsson Westergren, R, Stenmark, P. | | Deposit date: | 2022-09-25 | | Release date: | 2023-10-04 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional insights into the Pseudomonas aeruginosa glycosyltransferase WaaG and the implications for lipopolysaccharide biosynthesis.
J.Biol.Chem., 299, 2023
|
|
6B9X
 
 | | Crystal structure of Ragulator | | Descriptor: | Hepatitis B virus x interacting protein, Ragulator complex protein LAMTOR1, Ragulator complex protein LAMTOR2, ... | | Authors: | SU, M.-Y, Hurley, J.H. | | Deposit date: | 2017-10-11 | | Release date: | 2017-11-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Hybrid Structure of the RagA/C-Ragulator mTORC1 Activation Complex.
Mol. Cell, 68, 2017
|
|
6MQV
 
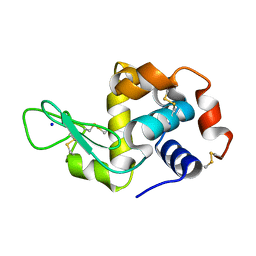 | | Structure of HEWL from LCP injector using synchrotron radiation | | Descriptor: | Lysozyme C, SODIUM ION | | Authors: | Caradoc-Davies, T.T, Aishima, J, Berntsen, P, Hadian-Jazi, M. | | Deposit date: | 2018-10-10 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The serial millisecond crystallography instrument at the Australian Synchrotron incorporating the "Lipidico" injector.
Rev Sci Instrum, 90, 2019
|
|
6CNI
 
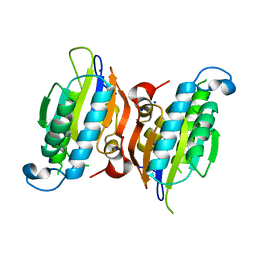 | | Crystal structure of H105A PGAM5 dimer | | Descriptor: | PHOSPHATE ION, SODIUM ION, Serine/threonine-protein phosphatase PGAM5, ... | | Authors: | Ruiz, K, Agnew, C, Jura, N. | | Deposit date: | 2018-03-08 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Functional role of PGAM5 multimeric assemblies and their polymerization into filaments.
Nat Commun, 10, 2019
|
|
8B6Z
 
 | |
8B6L
 
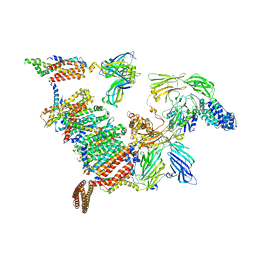 | | Subtomogram average of the human Sec61-TRAP-OSTA-translocon | | Descriptor: | Dolichyl-diphosphooligosaccharide--protein glycosyltransferase 48 kDa subunit, Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 1, Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 2, ... | | Authors: | Gemmer, M, Fedry, J.M.M, Forster, F.G. | | Deposit date: | 2022-09-27 | | Release date: | 2022-12-07 | | Last modified: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Visualization of translation and protein biogenesis at the ER membrane.
Nature, 614, 2023
|
|
5P2P
 
 | | X-RAY STRUCTURE OF PHOSPHOLIPASE A2 COMPLEXED WITH A SUBSTRATE-DERIVED INHIBITOR | | Descriptor: | CALCIUM ION, PHOSPHOLIPASE A2, PHOSPHONIC ACID 2-DODECANOYLAMINO-HEXYL ESTER PROPYL ESTER | | Authors: | Dijkstra, B.W, Thunnissen, M.M.G.M, Kalk, K.H, Drenth, J. | | Deposit date: | 1990-09-01 | | Release date: | 1991-10-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray structure of phospholipase A2 complexed with a substrate-derived inhibitor.
Nature, 347, 1990
|
|
7RLO
 
 | | Structure of the human eukaryotic translation initiation factor 2B (eIF2B) in complex with a viral protein NSs | | Descriptor: | Non-structural protein NS-S, Translation initiation factor eIF-2B subunit alpha, Translation initiation factor eIF-2B subunit beta, ... | | Authors: | Wang, L, Schoof, M, Cogan, J, Lawrence, R, Boone, M, Wuerth, J, Frost, M, Walter, P. | | Deposit date: | 2021-07-26 | | Release date: | 2021-12-22 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Viral evasion of the integrated stress response through antagonism of eIF2-P binding to eIF2B.
Nat Commun, 12, 2021
|
|
7A74
 
 | | Structure of G132N BlaC from Mycobacterium tuberculosis | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, GLYCEROL, ... | | Authors: | Chikunova, A, Ahmad, M.U, Perrakis, A, Ubbink, M. | | Deposit date: | 2020-08-27 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Two beta-Lactamase Variants with Reduced Clavulanic Acid Inhibition Display Different Millisecond Dynamics.
Antimicrob.Agents Chemother., 65, 2021
|
|
7R5T
 
 | | CRYSTAL STRUCTURE OF E.coli ALCOHOL DEHYDROGENASE - FucO MUTANT F254I COMPLEXED WITH FE, NADH, AND GLYCEROL | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ADENOSINE-5-DIPHOSPHORIBOSE, FE (III) ION, ... | | Authors: | Sridhar, S, Kiema, T.R, Wierenga, R, Widersten, M. | | Deposit date: | 2022-02-11 | | Release date: | 2022-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structures of lactaldehyde reductase, FucO, link enzyme activity to hydrogen bond networks and conformational dynamics.
Febs J., 290, 2023
|
|
7R3D
 
 | | CRYSTAL STRUCTURE OF E.coli ALCOHOL DEHYDROGENASE - FucO MUTANT N151G, L259V COMPLEXED WITH FE, NADH, AND GLYCEROL (Absence of Nicotinamide ring) | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, FE (III) ION, Lactaldehyde reductase | | Authors: | Sridhar, S, Kiema, T.R, Wierenga, R, Widersten, M. | | Deposit date: | 2022-02-07 | | Release date: | 2022-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structures of lactaldehyde reductase, FucO, link enzyme activity to hydrogen bond networks and conformational dynamics.
Febs J., 290, 2023
|
|
8BPP
 
 | |
8BPQ
 
 | |
