6GLC
 
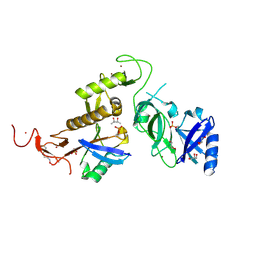 | | Structure of phospho-Parkin bound to phospho-ubiquitin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, E3 ubiquitin-protein ligase parkin, GLYCEROL, ... | | Authors: | Gladkova, C, Maslen, S.L, Skehel, J.M, Komander, D. | | Deposit date: | 2018-05-23 | | Release date: | 2018-06-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanism of parkin activation by PINK1.
Nature, 559, 2018
|
|
6HEM
 
 | |
6HEK
 
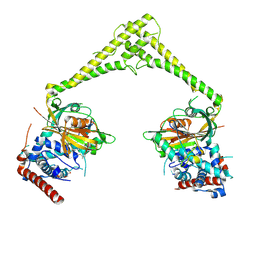 | | Structure of human USP28 bound to Ubiquitin-PA | | Descriptor: | CHLORIDE ION, Polyubiquitin-B, TETRAETHYLENE GLYCOL, ... | | Authors: | Gersch, M, Komander, D. | | Deposit date: | 2018-08-20 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Distinct USP25 and USP28 Oligomerization States Regulate Deubiquitinating Activity.
Mol.Cell, 74, 2019
|
|
4KVS
 
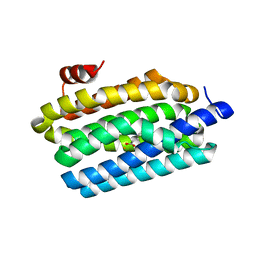 | | Crystal Structure of Prochlorococcus marinus aldehyde-deformylating oxygenase (mutant A134F) | | Descriptor: | Aldehyde decarbonylase, FE (III) ION, HEXANOIC ACID | | Authors: | Levy, C.W, Khara, B, Menon, N, Mansell, D, Das, D, Marsh, E.N.G, Leys, D, Scrutton, N.S. | | Deposit date: | 2013-05-23 | | Release date: | 2013-06-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Production of propane and other short-chain alkanes by structure-based engineering of ligand specificity in aldehyde-deformylating oxygenase.
Chembiochem, 14, 2013
|
|
1F27
 
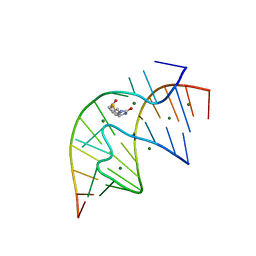 | | CRYSTAL STRUCTURE OF A BIOTIN-BINDING RNA PSEUDOKNOT | | Descriptor: | BIOTIN, MAGNESIUM ION, RNA (5'-R(*AP*AP*AP*AP*AP*GP*UP*CP*CP*UP*C)-3'), ... | | Authors: | Nix, J, Sussman, D, Wilson, C. | | Deposit date: | 2000-05-23 | | Release date: | 2000-06-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The 1.3 A crystal structure of a biotin-binding pseudoknot and the basis for RNA molecular recognition.
J.Mol.Biol., 296, 2000
|
|
8DI5
 
 | | Cryo-EM structure of SARS-CoV-2 Beta (B.1.351) spike protein in complex with VH domain F6 (focused refinement of RBD and VH F6) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, VH F6 | | Authors: | Zhu, X, Saville, J.W, Mannar, D, Berezuk, A.M, Subramaniam, S. | | Deposit date: | 2022-06-28 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Potent and broad neutralization of SARS-CoV-2 variants of concern (VOCs) including omicron sub-lineages BA.1 and BA.2 by biparatopic human VH domains.
Iscience, 25, 2022
|
|
8D9N
 
 | | CryoEM structures of bAE1 captured in multiple states. | | Descriptor: | Anion exchange protein | | Authors: | Zhekova, H.R, Wang, W.G, Jiang, J.S, Tsirulnikov, K, Muhammad-Khan, G.H, Azimov, R, Abuladze, N, Kao, L, Newman, D, Noskov, S.Y, Tieleman, P, Zhou, Z.H, Pushkin, A, Kurtz, I. | | Deposit date: | 2022-06-10 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | CryoEM structures of anion exchanger 1 capture multiple states of inward- and outward-facing conformations.
Commun Biol, 5, 2022
|
|
3F89
 
 | | NEMO CoZi domain | | Descriptor: | NF-kappa-B essential modulator | | Authors: | Rahighi, S, Ikeda, F, Kawasaki, M, Akutsu, M, Suzuki, N, Kato, R, Kensche, T, Uejima, T, Bloor, S, Komander, D, Randow, F, Wakatsuki, S, Dikic, I. | | Deposit date: | 2008-11-11 | | Release date: | 2009-03-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Specific recognition of linear ubiquitin chains by NEMO is important for NF-kappaB activation
Cell(Cambridge,Mass.), 136, 2009
|
|
8FU3
 
 | | Structure Of Respiratory Syncytial Virus Polymerase with Novel Non-Nucleoside Inhibitor | | Descriptor: | 8-methoxy-3-methyl-N-{(2S)-3,3,3-trifluoro-2-[5-fluoro-6-(4-fluorophenyl)-4-(2-hydroxypropan-2-yl)pyridin-2-yl]-2-hydroxypropyl}cinnoline-6-carboxamide, Phosphoprotein, RNA-directed RNA polymerase L | | Authors: | Yu, X, Abeywickrema, P, Bonneux, B, Behera, I, Jacoby, E, Fung, A, Adhikary, S, Bhaumik, A, Carbajo, R.J, Bruyn, S.D, Miller, R, Patrick, A, Pham, Q, Piassek, M, Verheyen, N, Shareef, A, Sutto-Ortiz, P, Ysebaert, N, Vlijmen, H.V, Jonckers, T.H.M, Herschke, F, McLellan, J.S, Decroly, E, Fearns, R, Grosse, S, Roymans, D, Sharma, S, Rigaux, P, Jin, Z. | | Deposit date: | 2023-01-16 | | Release date: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Structural and mechanistic insights into the inhibition of respiratory syncytial virus polymerase by a non-nucleoside inhibitor.
Commun Biol, 6, 2023
|
|
1H1W
 
 | | High resolution crystal structure of the human PDK1 catalytic domain | | Descriptor: | 3-PHOSPHOINOSITIDE DEPENDENT PROTEIN KINASE-1, ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, ... | | Authors: | Biondi, R.M, Komander, D, Thomas, C.C, Lizcano, J.M, Deak, M, Alessi, D.R, Van Aalten, D.M.F. | | Deposit date: | 2002-07-23 | | Release date: | 2003-07-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High Resolution Crystal Structure of the Human Pdk1 Catalytic Domain Defines the Regulatory Phosphopeptide Docking Site
Embo J., 21, 2003
|
|
8QVU
 
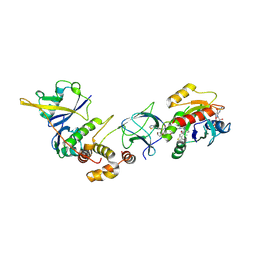 | | Crystal Structure of ligand ACBI3 in complex with KRAS G12D C118S GDP and pVHL:ElonginC:ElonginB complex | | Descriptor: | (2S,4R)-1-[(2S)-2-[4-[4-[(3S)-4-[4-[5-[(4S)-2-azanyl-3-cyano-4-methyl-6,7-dihydro-5H-1-benzothiophen-4-yl]-1,2,4-oxadiazol-3-yl]pyrimidin-2-yl]-3-methyl-1,4-diazepan-1-yl]butoxy]-1,2,3-triazol-1-yl]-3-methyl-butanoyl]-N-[(1R)-1-[4-(4-methyl-1,3-thiazol-5-yl)phenyl]-2-oxidanyl-ethyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Wijaya, A.J, Zollman, D, Farnaby, W, Ciulli, A. | | Deposit date: | 2023-10-18 | | Release date: | 2023-12-06 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Targeting cancer with small-molecule pan-KRAS degraders.
Science, 385, 2024
|
|
8DM2
 
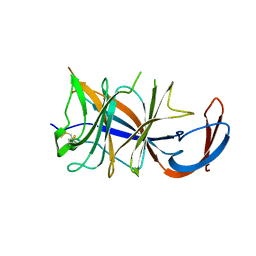 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2 spike protein (focused refinement of NTD) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, X, Saville, J.W, Mannar, D, Berezuk, A.M, Cholak, S, Tuttle, K.S, Vahdatihassani, F, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2023-01-25 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Structural analysis of receptor engagement and antigenic drift within the BA.2 spike protein.
Cell Rep, 42, 2023
|
|
8DM3
 
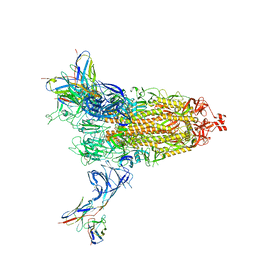 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2 spike protein in complex with Fab 4A8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 4A8 heavy chain, ... | | Authors: | Zhu, X, Saville, J.W, Mannar, D, Berezuk, A.M, Cholak, S, Tuttle, K.S, Vahdatihassani, F, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (2.37 Å) | | Cite: | Structural analysis of receptor engagement and antigenic drift within the BA.2 spike protein.
Cell Rep, 42, 2023
|
|
8DM1
 
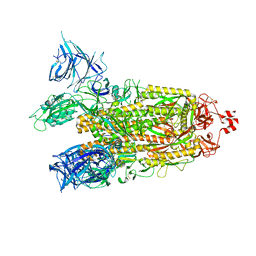 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2 spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, X, Saville, J.W, Mannar, D, Berezuk, A.M, Cholak, S, Tuttle, K.S, Vahdatihassani, F, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structural analysis of receptor engagement and antigenic drift within the BA.2 spike protein.
Cell Rep, 42, 2023
|
|
8DM4
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2 spike protein in complex with Fab 4A8 (focused refinement of NTD and 4A8) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 4A8 heavy chain, ... | | Authors: | Zhu, X, Saville, J.W, Mannar, D, Berezuk, A.M, Cholak, S, Tuttle, K.S, Vahdatihassani, F, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2023-01-25 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | Structural analysis of receptor engagement and antigenic drift within the BA.2 spike protein.
Cell Rep, 42, 2023
|
|
8E34
 
 | | CryoEM structures of bAE1 captured in multiple states | | Descriptor: | Anion exchange protein | | Authors: | Zhekova, H.R, Wang, W.G, Jiang, J.S, Tsirulnikov, K, Muhammad-Khan, G.H, Azimov, R, Abuladze, N, Kao, L, Newman, D, Noskov, S.Y, Tieleman, P, Zhou, Z.H, Pushkin, A, Kurtz, I. | | Deposit date: | 2022-08-16 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | CryoEM structures of anion exchanger 1 capture multiple states of inward- and outward-facing conformations.
Commun Biol, 5, 2022
|
|
8DM6
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2 spike protein in complex with human ACE2 (focused refinement of RBD and ACE2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Zhu, X, Saville, J.W, Mannar, D, Berezuk, A.M, Cholak, S, Tuttle, K.S, Vahdatihassani, F, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2023-02-08 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structural analysis of receptor engagement and antigenic drift within the BA.2 spike protein.
Cell Rep, 42, 2023
|
|
8DM8
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2 spike protein in complex with mouse ACE2 (focused refinement of RBD and ACE2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Zhu, X, Saville, J.W, Mannar, D, Berezuk, A.M, Cholak, S, Tuttle, K.S, Vahdatihassani, F, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2023-02-08 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Structural analysis of receptor engagement and antigenic drift within the BA.2 spike protein.
Cell Rep, 42, 2023
|
|
8DMA
 
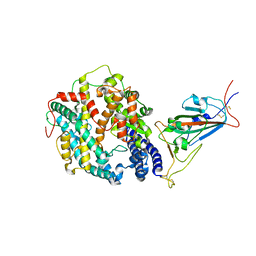 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.1 spike protein in complex with mouse ACE2 (focused refinement of RBD and ACE2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Zhu, X, Saville, J.W, Mannar, D, Berezuk, A.M, Cholak, S, Tuttle, K.S, Vahdatihassani, F, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Structural analysis of receptor engagement and antigenic drift within the BA.2 spike protein.
Cell Rep, 42, 2023
|
|
8DM7
 
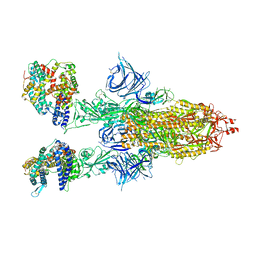 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2 spike protein in complex with mouse ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Saville, J.W, Mannar, D, Berezuk, A.M, Cholak, S, Tuttle, K.S, Vahdatihassani, F, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Structural analysis of receptor engagement and antigenic drift within the BA.2 spike protein.
Cell Rep, 42, 2023
|
|
8DM5
 
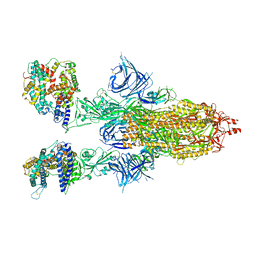 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2 spike protein in complex with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Saville, J.W, Mannar, D, Berezuk, A.M, Cholak, S, Tuttle, K.S, Vahdatihassani, F, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Structural analysis of receptor engagement and antigenic drift within the BA.2 spike protein.
Cell Rep, 42, 2023
|
|
8DM9
 
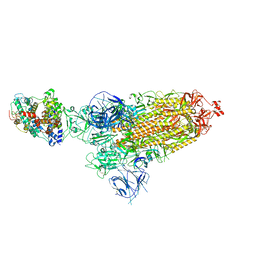 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.1 spike protein in complex with mouse ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Saville, J.W, Mannar, D, Berezuk, A.M, Cholak, S, Tuttle, K.S, Vahdatihassani, F, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Structural analysis of receptor engagement and antigenic drift within the BA.2 spike protein.
Cell Rep, 42, 2023
|
|
6M9T
 
 | | Crystal structure of EP3 receptor bound to misoprostol-FA | | Descriptor: | (11alpha,12alpha,13E,16S)-11,16-dihydroxy-16-methyl-9-oxoprost-13-en-1-oic acid, (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, OLEIC ACID, ... | | Authors: | Audet, M, White, K.L, Breton, B, Zarzycka, B, Han, G.W, Lu, Y, Gati, C, Batyuk, A, Popov, P, Velasquez, J, Manahan, D, Hu, H, Weierstall, U, Liu, W, Shui, W, Katrich, V, Cherezov, V, Hanson, M.A, Stevens, R.C. | | Deposit date: | 2018-08-24 | | Release date: | 2018-12-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of misoprostol bound to the labor inducer prostaglandin E2receptor.
Nat. Chem. Biol., 15, 2019
|
|
3EG5
 
 | | Crystal structure of MDIA1-TSH GBD-FH3 in complex with CDC42-GMPPNP | | Descriptor: | Cell division control protein 42 homolog, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Lammers, M, Meyer, S, Kuehlmann, D, Wittinghofer, A. | | Deposit date: | 2008-09-10 | | Release date: | 2008-10-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Specificity of Interactions between mDia Isoforms and Rho Proteins
J.Biol.Chem., 283, 2008
|
|
8EEQ
 
 | | CryoEM structures of bAE1 captured in multiple states. | | Descriptor: | Anion exchange protein | | Authors: | Zhekova, H.R, Wang, W.G, Jiang, J.S, Tsirulnikov, K, Muhammad-Khan, G.H, Azimov, R, Abuladze, N, Kao, L, Newman, D, Noskov, S.Y, Tieleman, P, Zhou, Z.H, Pushkin, A, Kurtz, I. | | Deposit date: | 2022-09-07 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | CryoEM structures of anion exchanger 1 capture multiple states of inward- and outward-facing conformations.
Commun Biol, 5, 2022
|
|
