5HDC
 
 | |
5HDD
 
 | |
5HDS
 
 | |
5BR1
 
 | | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM AGROBACTERIUM VITIS S4 (Avi_5305, TARGET EFI-511224) WITH BOUND ALPHA-D-GALACTOSAMINE | | Descriptor: | 2-amino-2-deoxy-alpha-D-galactopyranose, ABC transporter, binding protein | | Authors: | Yadava, U, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-05-29 | | Release date: | 2015-06-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of an ABC transporter solute-binding protein specific for the amino sugars glucosamine and galactosamine.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
3MTH
 
 | | X-RAY CRYSTALLOGRAPHIC STUDIES ON HEXAMERIC INSULINS IN THE PRESENCE OF HELIX-STABILIZING AGENTS, THIOCYANATE, METHYLPARABEN AND PHENOL | | Descriptor: | 4-HYDROXY-BENZOIC ACID METHYL ESTER, CHLORIDE ION, METHYLPARABEN INSULIN, ... | | Authors: | Whittingham, J.L, Dodson, E.J, Moody, P.C.E, Dodson, G.G. | | Deposit date: | 1995-09-13 | | Release date: | 1996-01-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray crystallographic studies on hexameric insulins in the presence of helix-stabilizing agents, thiocyanate, methylparaben, and phenol.
Biochemistry, 34, 1995
|
|
4Z8K
 
 | |
6ELC
 
 | | Crystal Structure of O-linked Glycosylated VSG3 | | Descriptor: | Variant surface glycoprotein, alpha-D-glucopyranose, alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Stebbins, C.E. | | Deposit date: | 2017-09-28 | | Release date: | 2018-07-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | African trypanosomes evade immune clearance by O-glycosylation of the VSG surface coat.
Nat Microbiol, 3, 2018
|
|
5VXV
 
 | | Peroxisomal membrane protein PEX15 | | Descriptor: | Peroxisomal membrane protein PEX15 | | Authors: | Gardner, B.M, Castanzo, D.T. | | Deposit date: | 2017-05-24 | | Release date: | 2018-01-17 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The peroxisomal AAA-ATPase Pex1/Pex6 unfolds substrates by processive threading.
Nat Commun, 9, 2018
|
|
5V68
 
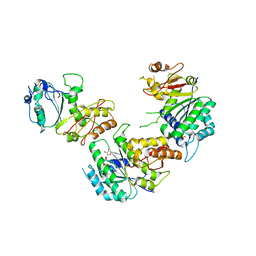 | | Crystal structure of cell division protein FtsZ from Mycobacterium tuberculosis bounded via the T9 loop | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, PHOSPHATE ION | | Authors: | Lazo, E.O, Ojima, I, Chowdhury, S.R, Awasthi, D, Jakoncic, J. | | Deposit date: | 2017-03-16 | | Release date: | 2017-03-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | Novel T9 loop conformation of filamenting temperature-sensitive mutant Z from Mycobacterium tuberculosis.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6XFV
 
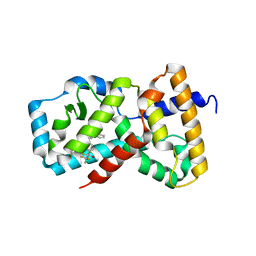 | | CRYSTAL STRUCTURE OF RAR-RELATED ORPHAN RECEPTOR C IN COMPLEX WITH A NOVEL INVERSE AGONIST | | Descriptor: | 1-(4-{(3S,4S)-4-[4-(1,1,1,3,3,3-hexafluoro-2-hydroxypropan-2-yl)phenyl]-3-methyl-3-phenylpyrrolidine-1-carbonyl}piperidin-1-yl)ethan-1-one, Nuclear receptor ROR-gamma | | Authors: | Sack, J.S. | | Deposit date: | 2020-06-16 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of (3S,4S)-3-methyl-3-(4-fluorophenyl)-4-(4-(1,1,1,3,3,3-hexafluoro-2-hydroxyprop-2-yl)phenyl)pyrrolidines as novel ROR gamma t inverse agonists.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
7V3W
 
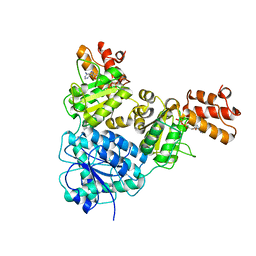 | | Crystal Structure of VpsR display novel dimeric architecture and c-di-GMP binding: mechanistic implications in oligomerization, ATPase activity and DNA binding. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, VpsR | | Authors: | Chakrabortty, T, Sen, U, Chowdhury, S.R. | | Deposit date: | 2021-08-11 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.205 Å) | | Cite: | Crystal Structure of VpsR Revealed Novel Dimeric Architecture and c-di-GMP Binding Site: Mechanistic Implications in Oligomerization, ATPase Activity and DNA Binding.
J.Mol.Biol., 434, 2022
|
|
7V2V
 
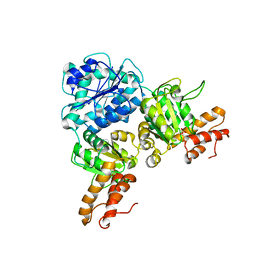 | | Crystal Structure of VpsR display novel dimeric architecture and c-di-GMP binding: mechanistic implications in oligomerization, ATPase activity and DNA binding. | | Descriptor: | SULFATE ION, VpsR | | Authors: | Chakrabortty, T, Sen, U, Chowdhury, S.R. | | Deposit date: | 2021-08-10 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.194 Å) | | Cite: | Crystal Structure of VpsR Revealed Novel Dimeric Architecture and c-di-GMP Binding Site: Mechanistic Implications in Oligomerization, ATPase Activity and DNA Binding.
J.Mol.Biol., 434, 2022
|
|
6NZ7
 
 | |
7UEB
 
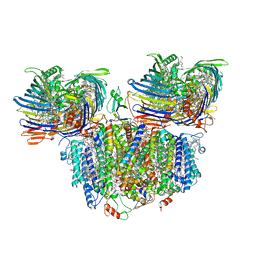 | | Photosynthetic assembly of Chlorobaculum tepidum (RC-FMO2) | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, 2-[(1E,3E,5E,7E,9E,11E,13E,15E,17E,19E)-3,7,12,16,20,24-hexamethylpentacosa-1,3,5,7,9,11,13,15,17,19,23-undecaenyl]-1,3,4-trimethyl-benzene, ... | | Authors: | Puskar, R, Truong, C.D, Swain, K, Li, S, Cheng, K.-W, Wang, T.Y, Poh, Y.-P, Liu, H, Chou, T.-F, Nannenga, B, Chiu, P.-L. | | Deposit date: | 2022-03-21 | | Release date: | 2022-10-05 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Molecular asymmetry of a photosynthetic supercomplex from green sulfur bacteria.
Nat Commun, 13, 2022
|
|
7UEA
 
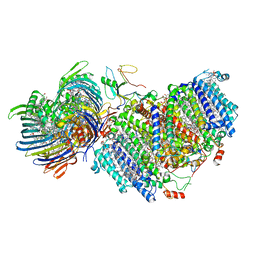 | | Photosynthetic assembly of Chlorobaculum tepidum (RC-FMO1) | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, 2-[(1E,3E,5E,7E,9E,11E,13E,15E,17E,19E)-3,7,12,16,20,24-hexamethylpentacosa-1,3,5,7,9,11,13,15,17,19,23-undecaenyl]-1,3,4-trimethyl-benzene, ... | | Authors: | Puskar, R, Truong, C.D, Swain, K, Li, S, Cheng, K.-W, Wang, T.Y, Poh, Y.-P, Liu, H, Chou, T.-F, Nannenga, B, Chiu, P.-L. | | Deposit date: | 2022-03-21 | | Release date: | 2022-10-05 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Molecular asymmetry of a photosynthetic supercomplex from green sulfur bacteria.
Nat Commun, 13, 2022
|
|
4S0H
 
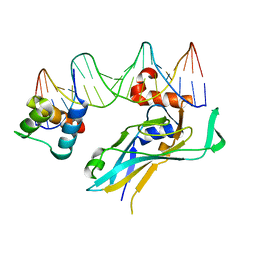 | | TBX5 DB, NKX2.5 HD, ANF DNA Complex | | Descriptor: | 5'-D(*CP*CP*AP*CP*TP*TP*CP*AP*AP*AP*GP*GP*TP*GP*TP*GP*AP*GP*A)-3', 5'-D(*TP*CP*TP*CP*AP*CP*AP*CP*CP*TP*TP*TP*GP*AP*AP*GP*TP*GP*G)-3', Homeobox protein Nkx-2.5, ... | | Authors: | Pradhan, L. | | Deposit date: | 2014-12-31 | | Release date: | 2015-12-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.817 Å) | | Cite: | Intermolecular Interactions of Cardiac Transcription Factors NKX2.5 and TBX5.
Biochemistry, 55, 2016
|
|
7UPX
 
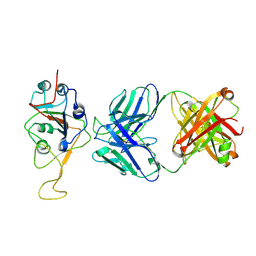 | | Three RBD-down state of SARS-CoV-2 D614G spike in complex with the SP1-77 neutralizing antibody Fab fragment (local refinement of the RBD and Fab variable domains) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, SP1-77 Fab heavy chain, SP1-77 Fab light chain, ... | | Authors: | Zhang, J, Luo, S, Kreutzberger, A, Kirchhausen, T, Chen, B, Haynes, B, Alt, F. | | Deposit date: | 2022-04-18 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | An antibody from single human V H -rearranging mouse neutralizes all SARS-CoV-2 variants through BA.5 by inhibiting membrane fusion.
Sci Immunol, 7, 2022
|
|
7UPY
 
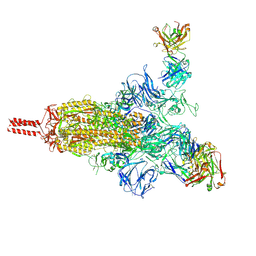 | | An antibody from single human VH-rearranging mouse neutralizes all SARS-CoV-2 variants through BA.5 by inhibiting membrane fusion | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Luo, S, Kreutzberger, A, Kirchhausen, T, Chen, B, Haynes, B, Alt, F. | | Deposit date: | 2022-04-18 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | An antibody from single human V H -rearranging mouse neutralizes all SARS-CoV-2 variants through BA.5 by inhibiting membrane fusion.
Sci Immunol, 7, 2022
|
|
7UPW
 
 | | Three RBD-down state of SARS-CoV-2 D614G spike in complex with the SP1-77 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Luo, S, Kreutzberger, A, Kirchhausen, T, Chen, B, Haynes, B, Alt, F. | | Deposit date: | 2022-04-18 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | An antibody from single human V H -rearranging mouse neutralizes all SARS-CoV-2 variants through BA.5 by inhibiting membrane fusion.
Sci Immunol, 7, 2022
|
|
6O3T
 
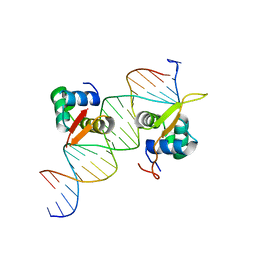 | | Structural basis of FOXC2 and DNA interactions | | Descriptor: | DNA (5'-D(*AP*AP*AP*TP*TP*GP*TP*TP*TP*AP*TP*AP*AP*AP*CP*AP*GP*CP*CP*CP*G)-3'), DNA (5'-D(*TP*TP*CP*GP*GP*GP*CP*TP*GP*TP*TP*TP*AP*TP*AP*AP*AP*CP*AP*AP*T)-3'), Forkhead box protein C2 | | Authors: | Nam, H.-J, Li, S. | | Deposit date: | 2019-02-27 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Crystal Structure of FOXC2 in Complex with DNA Target.
Acs Omega, 4, 2019
|
|
6S41
 
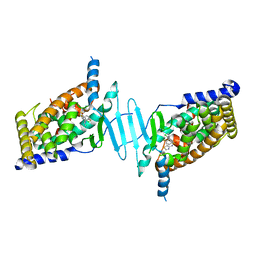 | | CRYSTAL STRUCTURE OF PXR IN COMPLEX WITH XPC-7455 | | Descriptor: | 4-[[(1~{S})-1-[2,5-bis(fluoranyl)phenyl]ethyl]amino]-5-chloranyl-2-fluoranyl-~{N}-(1,3-thiazol-4-yl)benzenesulfonamide, Nuclear receptor subfamily 1 group I member 2 | | Authors: | Focken, T, Maskos, K, Griessner, A, Krapp, S. | | Deposit date: | 2019-06-26 | | Release date: | 2019-10-02 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identification of CNS-Penetrant Aryl Sulfonamides as Isoform-Selective NaV1.6 Inhibitors with Efficacy in Mouse Models of Epilepsy.
J.Med.Chem., 62, 2019
|
|
7SG4
 
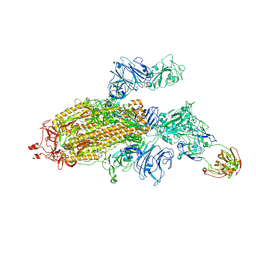 | |
5VO6
 
 | |
8GB6
 
 | |
8GB7
 
 | |
