1NXB
 
 | |
1IGN
 
 | | DNA-BINDING DOMAIN OF RAP1 IN COMPLEX WITH TELOMERIC DNA SITE | | Descriptor: | DNA (5'-D(*CP*CP*GP*CP*AP*CP*AP*CP*CP*CP*AP*CP*AP*CP*AP*CP*C P*AP*G)-3'), DNA (5'-D(*CP*CP*TP*GP*GP*TP*GP*TP*GP*TP*GP*GP*GP*TP*GP*TP*G P*CP*G)-3'), PROTEIN (RAP1) | | Authors: | Koenig, P, Giraldo, R, Chapman, L, Rhodes, D. | | Deposit date: | 1996-02-29 | | Release date: | 1997-01-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The crystal structure of the DNA-binding domain of yeast RAP1 in complex with telomeric DNA.
Cell(Cambridge,Mass.), 85, 1996
|
|
5LPM
 
 | | Crystal structure of the bromodomain of human Ep300 bound to the inhibitor XDM3d | | Descriptor: | ACETATE ION, Histone acetyltransferase p300, ~{N}-[(1~{S},2~{S})-7-chloranyl-2-oxidanyl-1,2,3,4-tetrahydronaphthalen-1-yl]-4-ethanoyl-3-ethyl-5-methyl-1~{H}-pyrrole -2-carboxamide | | Authors: | Huegle, M, Wohlwend, D, Gerhardt, S. | | Deposit date: | 2016-08-13 | | Release date: | 2017-08-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Beyond the BET Family: Targeting CBP/p300 with 4-Acyl Pyrroles.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
6O2R
 
 | | Deacetylated Microtubules | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Eshun-Wilson, L, Zhang, R, Portran, D, Nachury, M.V, Toso, D, Lohr, T, Vendruscolo, M, Bonomi, M, Fraser, J.S, Nogales, E. | | Deposit date: | 2019-02-24 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Effects of alpha-tubulin acetylation on microtubule structure and stability.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5AP8
 
 | | Structure of the SAM-dependent rRNA:acp-transferase Tsr3 from S. solfataricus | | Descriptor: | TSR3 | | Authors: | Wurm, J.P, Immer, C, Pogoryelov, D, Meyer, B, Koetter, P, Entian, K.-D, Woehnert, J. | | Deposit date: | 2015-09-14 | | Release date: | 2016-04-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.246 Å) | | Cite: | Ribosome Biogenesis Factor Tsr3 is the Aminocarboxypropyl Transferase Responsible for 18S Rrna Hypermodification in Yeast and Humans
Nucleic Acids Res., 44, 2016
|
|
3OXU
 
 | | Complement components factor H CCP19-20 and C3d in complex | | Descriptor: | Complement C3, GLYCEROL, HF protein | | Authors: | Morgan, H.P, Schmidt, C.Q, Guariento, M, Gillespie, D, Herbert, A.P, Mertens, H, Blaum, B.S, Svergun, D, Johansson, C.M, Uhrin, D, Barlow, P.N, Hannan, J.P. | | Deposit date: | 2010-09-22 | | Release date: | 2011-02-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for engagement by complement factor H of C3b on a self surface.
Nat.Struct.Mol.Biol., 18, 2011
|
|
6J2J
 
 | | Crystal structure of bat (Pteropus Alecto) MHC class I Ptal-N*01:01 in complex with MERS-CoV-derived peptide MERS-CoV-S3 | | Descriptor: | Beta-2-microglobulin, MERS-CoV-S3, Ptal-N*01:01 | | Authors: | Lu, D, Liu, K.F, Yue, C, Lu, Q, Cheng, H, Chai, Y, Qi, J.X, Gao, G.F, Liu, W.J. | | Deposit date: | 2019-01-01 | | Release date: | 2019-09-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Peptide presentation by bat MHC class I provides new insight into the antiviral immunity of bats.
Plos Biol., 17, 2019
|
|
1NX2
 
 | | Calpain Domain VI | | Descriptor: | CALCIUM ION, Calcium-dependent protease, small subunit | | Authors: | Todd, B, Moore, D, Deivanayagam, C.C.S, Lin, G.-D, Chattopadhyay, D, Maki, M, Wang, K.K.W, Narayana, S.V.L. | | Deposit date: | 2003-02-07 | | Release date: | 2003-08-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A structural model for the inhibition of calpain by calpastatin: crystal structures of the native domain VI of calpain and its complexes with calpastatin peptide and a small molecule inhibitor.
J.Mol.Biol., 328, 2003
|
|
1AU6
 
 | | SOLUTION STRUCTURE OF DNA D(CATGCATG) INTERSTRAND-CROSSLINKED BY BISPLATIN COMPOUND (1,1/T,T), NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | BIS(TRANS-PLATINUM ETHYLENEDIAMINE DIAMINE CHLORO)COMPLEX, DNA (5'-D(*CP*AP*TP*GP*CP*AP*TP*G)-3') | | Authors: | Yang, D, Van Boom, S.S.G.E, Reedijk, J, Van Boom, J.H, Farrell, N, Wang, A.H.-J. | | Deposit date: | 1997-09-11 | | Release date: | 1998-02-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A novel DNA structure induced by the anticancer bisplatinum compound crosslinked to a GpC site in DNA.
Nat.Struct.Biol., 2, 1995
|
|
1NYH
 
 | | Crystal Structure of the Coiled-coil Dimerization Motif of Sir4 | | Descriptor: | Regulatory protein SIR4 | | Authors: | Chang, J.F, Hall, B.E, Tanny, J.C, Moazed, D, Filman, D, Ellenberger, T. | | Deposit date: | 2003-02-12 | | Release date: | 2003-06-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the Coiled-coil Dimerization Motif of Sir4 and Its Interaction With Sir3
Structure, 11, 2003
|
|
7W5N
 
 | | The crystal structure of the reduced form of Gluconobacter oxydans WSH-004 SNDH | | Descriptor: | L-sorbosone dehydrogenase, NAD(P) dependent, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Li, D, Hou, X.D, Rao, Y.J, Yin, D.J, Zhou, J.W, Chen, J. | | Deposit date: | 2021-11-30 | | Release date: | 2023-03-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.988 Å) | | Cite: | Structural Insight into the Catalytic Mechanisms of an L-Sorbosone Dehydrogenase.
Adv Sci, 10, 2023
|
|
7W5K
 
 | | The C296A mutant of L-sorbosone dehydrogenase (SNDH) from Gluconobacter Oxydans WSH-004 | | Descriptor: | L-sorbosone dehydrogenase, NAD(P) dependent, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Li, D, Hou, X.D, Rao, Y.J, Zhou, J.W, Chen, J. | | Deposit date: | 2021-11-30 | | Release date: | 2023-03-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural Insight into the Catalytic Mechanisms of an L-Sorbosone Dehydrogenase.
Adv Sci, 10, 2023
|
|
1K1C
 
 | | Solution Structure of Crh, the Bacillus subtilis Catabolite Repression HPr | | Descriptor: | catabolite repression HPr-like protein | | Authors: | Favier, A, Brutscher, B, Blackledge, M, Galinier, A, Deutscher, J, Penin, F, Marion, D. | | Deposit date: | 2001-09-25 | | Release date: | 2001-10-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of Crh, the Bacillus subtilis catabolite repression HPr.
J.Mol.Biol., 317, 2002
|
|
5LPL
 
 | | Crystal structure of the bromodomain of human CREBBP bound to the inhibitor XDM3c | | Descriptor: | CREB-binding protein, ~{N}-[(1~{R},2~{R})-7-chloranyl-2-oxidanyl-1,2,3,4-tetrahydronaphthalen-1-yl]-4-ethanoyl-3-ethyl-5-methyl-1~{H}-pyrrole-2-carboxamide | | Authors: | Wohlwend, D, Huegle, M. | | Deposit date: | 2016-08-13 | | Release date: | 2017-08-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Beyond the BET Family: Targeting CBP/p300 with 4-Acyl Pyrroles.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
1JPS
 
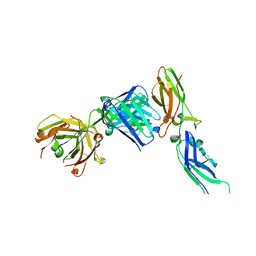 | | Crystal structure of tissue factor in complex with humanized Fab D3h44 | | Descriptor: | immunoglobulin Fab D3H44, heavy chain, light chain, ... | | Authors: | Faelber, K, Kirchhofer, D, Presta, L, Kelley, R.F, Muller, Y.A. | | Deposit date: | 2001-08-03 | | Release date: | 2002-02-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The 1.85 A resolution crystal structures of tissue factor in complex with humanized Fab D3h44 and of free humanized Fab D3h44: revisiting the solvation of antigen combining sites.
J.Mol.Biol., 313, 2001
|
|
5NCC
 
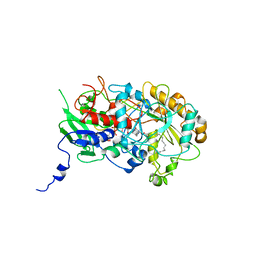 | | Structure of Fatty acid Photodecarboxylase in complex with FAD and palmitic acid | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid Photodecarboxylase, PALMITIC ACID | | Authors: | Arnoux, P, Sorigue, D, Beisson, F, Pignol, D. | | Deposit date: | 2017-03-03 | | Release date: | 2017-08-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | An algal photoenzyme converts fatty acids to hydrocarbons.
Science, 357, 2017
|
|
1JPT
 
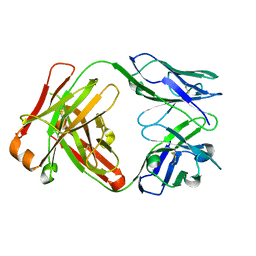 | | Crystal Structure of Fab D3H44 | | Descriptor: | immunoglobulin Fab D3H44, heavy chain, immunoglobulin Fab D3h44, ... | | Authors: | Faelber, K, Kirchhofer, D, Presta, L, Kelley, R.F, Muller, Y.A. | | Deposit date: | 2001-08-03 | | Release date: | 2002-02-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The 1.85 A resolution crystal structures of tissue factor in complex with humanized Fab D3h44 and of free humanized Fab D3h44: revisiting the solvation of antigen combining sites.
J.Mol.Biol., 313, 2001
|
|
5M63
 
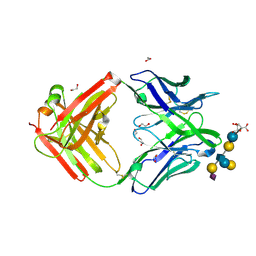 | | Crystal structure of group B Streptococcus type III DP2 oligosaccharide bound to Fab NVS-1-19-5 | | Descriptor: | (2~{R},3~{R},4~{S},5~{S})-2-[bis(oxidanyl)methyl]-5-(hydroxymethyl)oxolane-3,4-diol, 1,2-ETHANEDIOL, H chain of Fab NVS-1-19-5, ... | | Authors: | Carboni, F, Adamo, R, Veggi, D, Rappuoli, R, Malito, E, Margarit, I.R, Berti, F. | | Deposit date: | 2016-10-24 | | Release date: | 2017-05-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structure of a protective epitope of group B Streptococcus type III capsular polysaccharide.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5MJP
 
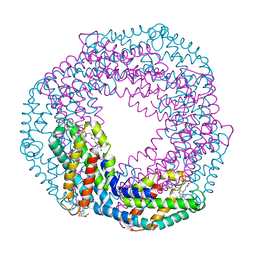 | | Multi-bunch pink beam serial crystallography: Phycocyanin (One chip) | | Descriptor: | C-phycocyanin alpha chain, C-phycocyanin beta chain, PHYCOCYANOBILIN | | Authors: | Meents, A, Oberthuer, D, Lieske, J, Srajer, V, Sarrou, I. | | Deposit date: | 2016-12-01 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Pink-beam serial crystallography.
Nat Commun, 8, 2017
|
|
1BD0
 
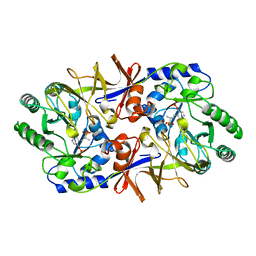 | | ALANINE RACEMASE COMPLEXED WITH ALANINE PHOSPHONATE | | Descriptor: | ALANINE RACEMASE, {1-[(3-HYDROXY-METHYL-5-PHOSPHONOOXY-METHYL-PYRIDIN-4-YLMETHYL)-AMINO]-ETHYL}-PHOSPHONIC ACID | | Authors: | Stamper, G.F, Morollo, A.A, Ringe, D. | | Deposit date: | 1998-05-12 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Reaction of alanine racemase with 1-aminoethylphosphonic acid forms a stable external aldimine.
Biochemistry, 37, 1998
|
|
1SGK
 
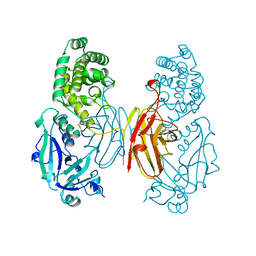 | | NUCLEOTIDE-FREE DIPHTHERIA TOXIN | | Descriptor: | DIPHTHERIA TOXIN (DIMERIC) | | Authors: | Bell, C.E, Eisenberg, D. | | Deposit date: | 1996-09-12 | | Release date: | 1996-12-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of nucleotide-free diphtheria toxin.
Biochemistry, 36, 1997
|
|
7WJU
 
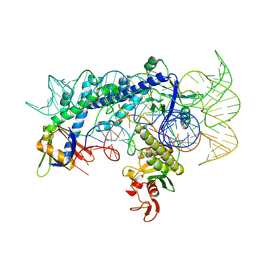 | | Cryo-EM structure of the AsCas12f1-sgRNAv1-dsDNA ternary complex | | Descriptor: | Non-target strand, OrfB_Zn_ribbon domain-containing protein, Target strand, ... | | Authors: | Wu, Z, Liu, D, Shen, H, Ji, Q. | | Deposit date: | 2022-01-07 | | Release date: | 2023-01-18 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Structure-directed functional evolution of the miniature CRISPR-AsCas12f1 system
To Be Published
|
|
1AU5
 
 | | SOLUTION STRUCTURE OF INTRASTRAND CISPLATIN-CROSSLINKED DNA OCTAMER D(CCTG*G*TCC):D(GGACCAGG), NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | Cisplatin, DNA (5'-D(*CP*CP*TP*GP*GP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*CP*CP*AP*GP*G)-3') | | Authors: | Yang, D, Van Boom, S.S.G.E, Reedijk, J, Van Boom, J.H, Wang, A.H.-J. | | Deposit date: | 1997-09-11 | | Release date: | 1998-02-25 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Structure and isomerization of an intrastrand cisplatin-cross-linked octamer DNA duplex by NMR analysis.
Biochemistry, 34, 1995
|
|
7WQV
 
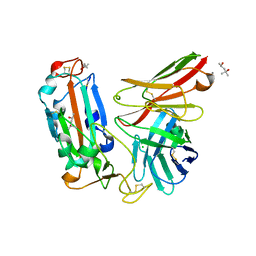 | | Crystal structure of a neutralizing monoclonal antibody (Ab08) in complex with SARS-CoV-2 receptor-binding domain (RBD) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Ab08, ... | | Authors: | Zha, J, Meng, L, Zhang, X, Li, D. | | Deposit date: | 2022-01-26 | | Release date: | 2023-01-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Spike-destructing human antibody effectively neutralizes Omicron-included SARS-CoV-2 variants with therapeutic efficacy.
Plos Pathog., 19, 2023
|
|
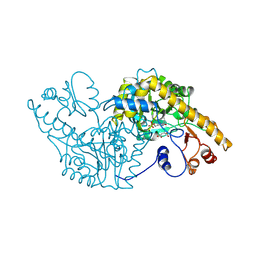
1ASE
 
 | |
