7VNC
 
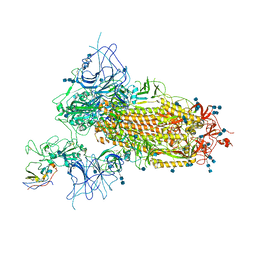 | | Structure of the SARS-CoV-2 spike glycoprotein in complex with a human single domain antibody n3113 (UDD-state, state 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yang, Z, Wang, Y, Kong, Y, Jin, Y, Wu, Y, Ying, T. | | Deposit date: | 2021-10-10 | | Release date: | 2021-11-24 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | A non-ACE2 competing human single-domain antibody confers broad neutralization against SARS-CoV-2 and circulating variants.
Signal Transduct Target Ther, 6, 2021
|
|
7XVR
 
 | |
7XVV
 
 | |
7XVW
 
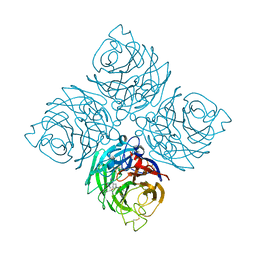 | | Structure of neuraminidase from influenza B-like viruses derived from spiny eel | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(1-ACETYLAMINO-2-ETHYL-BUTYL)-4-GUANIDINO-2-HYDROXY-CYCLOPENTANECARBOXYLIC ACID, CALCIUM ION, ... | | Authors: | Chai, Y, Gao, F. | | Deposit date: | 2022-05-25 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural and inhibitor sensitivity analysis of influenza B-like viral neuraminidases derived from Asiatic toad and spiny eel.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XVU
 
 | | Structure of neuraminidase from influenza B-like viruses derived from spiny eel | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Chai, Y, Gao, F. | | Deposit date: | 2022-05-24 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and inhibitor sensitivity analysis of influenza B-like viral neuraminidases derived from Asiatic toad and spiny eel.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7V5H
 
 | |
7XGR
 
 | |
7XDT
 
 | |
7Y4F
 
 | | bacterial DPP4 | | Descriptor: | Dipeptidyl peptidase IV | | Authors: | Hang, J, Jiang, C, Wang, K, Zhang, Z, Guo, F, Liu, J, Wang, G, Lei, X, Gonzalez, F, Qiao, J. | | Deposit date: | 2022-06-14 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.918 Å) | | Cite: | Microbial-host-isozyme analyses reveal microbial DPP4 as a potential antidiabetic target.
Science, 381, 2023
|
|
7Y4G
 
 | | sit-bound btDPP4 | | Descriptor: | (2R)-4-OXO-4-[3-(TRIFLUOROMETHYL)-5,6-DIHYDRO[1,2,4]TRIAZOLO[4,3-A]PYRAZIN-7(8H)-YL]-1-(2,4,5-TRIFLUOROPHENYL)BUTAN-2-A MINE, btDPP4 | | Authors: | Hang, J, Jiang, C, Wang, K, Zhang, Z, Guo, F, Liu, J, Wang, G, Lei, X, Gonzalez, F, Qiao, J. | | Deposit date: | 2022-06-14 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Microbial-host-isozyme analyses reveal microbial DPP4 as a potential antidiabetic target.
Science, 381, 2023
|
|
3DBS
 
 | | Structure of PI3K gamma in complex with GDC0941 | | Descriptor: | 2-(1H-indazol-4-yl)-6-{[4-(methylsulfonyl)piperazin-1-yl]methyl}-4-morpholin-4-yl-thieno[3,2-d]pyrimidine, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Wiesmann, C, Ultsch, M. | | Deposit date: | 2008-06-02 | | Release date: | 2008-06-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The identification of 2-(1H-indazol-4-yl)-6-(4-methanesulfonyl-piperazin-1-ylmethyl)-4-morpholin-4-yl-thieno[3,2-d]pyrimidine (GDC-0941) as a potent, selective, orally bioavailable inhibitor of class I PI3 kinase for the treatment of cancer
J.Med.Chem., 51, 2008
|
|
7YD1
 
 | |
7YD0
 
 | |
7YCY
 
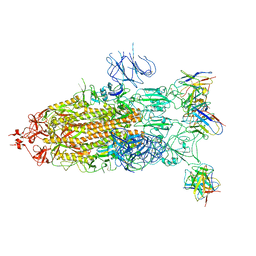 | |
7YCZ
 
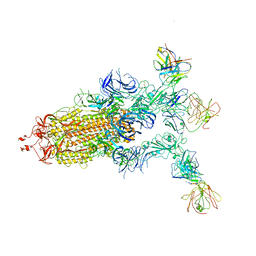 | |
6XT7
 
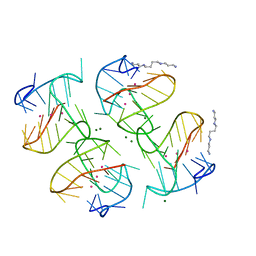 | | Tel25 Hybrid Four-quartet G-quadruplex with K+ | | Descriptor: | DNA (25-MER), MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Yatsunyk, L.A, McCarthy, S.E. | | Deposit date: | 2020-07-17 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | The first crystal structures of hybrid and parallel four-tetrad intramolecular G-quadruplexes.
Nucleic Acids Res., 50, 2022
|
|
7KLP
 
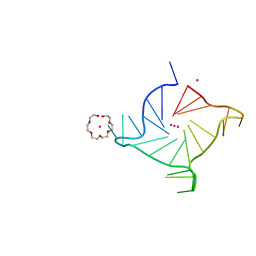 | | Crystal structure of a three-tetrad, parallel, K+ stabilized human telomeric G-quadruplex | | Descriptor: | HEXAETHYLENE GLYCOL, POTASSIUM ION, telomeric DNA | | Authors: | Yatsunyk, L.A, Li, K.S, Chen, E.V. | | Deposit date: | 2020-10-30 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Water spines and networks in G-quadruplex structures.
Nucleic Acids Res., 49, 2021
|
|
7RC9
 
 | | Crystal structure of human TLR8 ectodomain bound to small molecule antagonist 21 | | Descriptor: | 2-(2,6-dimethylpyridin-4-yl)-5-(piperidin-4-yl)-3-(propan-2-yl)-1H-indole, 2-acetamido-2-deoxy-beta-D-glucopyranose, Toll-like receptor 8, ... | | Authors: | Critton, D.A. | | Deposit date: | 2021-07-07 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Identification of 2-Pyridinylindole-Based Dual Antagonists of Toll-like Receptors 7 and 8 (TLR7/8).
Acs Med.Chem.Lett., 13, 2022
|
|
4EPU
 
 | | Ang1 fibrinogen-related domain (FReD) | | Descriptor: | Angiopoietin-1, CALCIUM ION | | Authors: | Yeykal, C.C, Adams, E.J. | | Deposit date: | 2012-04-17 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | Biochemical and structural characterization of the Ang1 fibrinogen-related domain
To be Published
|
|
6V9U
 
 | | Crystal structure of human TLR8 ectodomain bound to small molecule antagonist 14c | | Descriptor: | 2-(3,4-dimethoxyphenyl)-5-(piperidin-4-yl)-3-(propan-2-yl)-1H-indole, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Critton, D.A. | | Deposit date: | 2019-12-16 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Discovery of Potent and Orally Bioavailable Small Molecule Antagonists of Toll-like Receptors 7/8/9 (TLR7/8/9).
Acs Med.Chem.Lett., 11, 2020
|
|
5CT7
 
 | | BRAF in Complex with RAF265 | | Descriptor: | 1-methyl-5-({2-[5-(trifluoromethyl)-1H-imidazol-2-yl]pyridin-4-yl}oxy)-N-[4-(trifluoromethyl)phenyl]-1H-benzimidazol-2-amine, Serine/threonine-protein kinase B-raf | | Authors: | Appleton, B.A. | | Deposit date: | 2015-07-23 | | Release date: | 2015-09-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Discovery of RAF265: A Potent mut-B-RAF Inhibitor for the Treatment of Metastatic Melanoma.
Acs Med.Chem.Lett., 6, 2015
|
|
