1RZ1
 
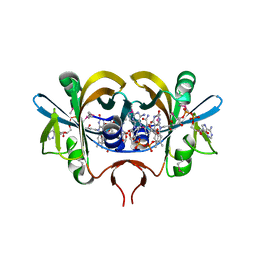 | | Reduced flavin reductase PheA2 in complex with NAD | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, phenol 2-hydroxylase component B | | Authors: | Van Den Heuvel, R.H, Westphal, A.H, Heck, A.J, Walsh, M.A, Rovida, S, Van Berkel, W.J, Mattevi, A. | | Deposit date: | 2003-12-23 | | Release date: | 2004-04-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Studies on Flavin Reductase PheA2 Reveal Binding of NAD in an Unusual Folded Conformation and Support Novel Mechanism of Action.
J.Biol.Chem., 279, 2004
|
|
6R1T
 
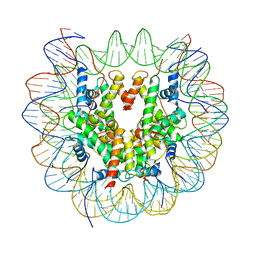 | | Structure of LSD2/NPAC-linker/nucleosome core particle complex: Class 1, free nuclesome | | Descriptor: | DNA (147-MER), HISTONE H2A, Histone H2A, ... | | Authors: | Marabelli, C, Pilotto, S, Chittori, S, Subramaniam, S, Mattevi, A. | | Deposit date: | 2019-03-15 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.02 Å) | | Cite: | A Tail-Based Mechanism Drives Nucleosome Demethylation by the LSD2/NPAC Multimeric Complex.
Cell Rep, 27, 2019
|
|
6R1U
 
 | | Structure of LSD2/NPAC-linker/nucleosome core particle complex: Class 2 | | Descriptor: | DNA (147-MER), FLAVIN-ADENINE DINUCLEOTIDE, Histone H2A, ... | | Authors: | Marabelli, C, Pilotto, S, Chittori, S, Subramaniam, S, Mattevi, A. | | Deposit date: | 2019-03-15 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.36 Å) | | Cite: | A Tail-Based Mechanism Drives Nucleosome Demethylation by the LSD2/NPAC Multimeric Complex.
Cell Rep, 27, 2019
|
|
6R25
 
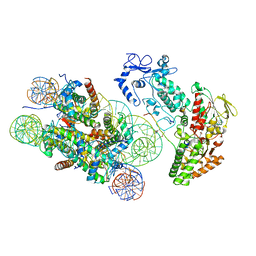 | | Structure of LSD2/NPAC-linker/nucleosome core particle complex: Class 3 | | Descriptor: | DNA (147-MER), FLAVIN-ADENINE DINUCLEOTIDE, H2B, ... | | Authors: | Marabelli, C, Pilotto, S, Chittori, S, Subramaniam, S, Mattevi, A. | | Deposit date: | 2019-03-15 | | Release date: | 2019-04-24 | | Method: | ELECTRON MICROSCOPY (4.61 Å) | | Cite: | A Tail-Based Mechanism Drives Nucleosome Demethylation by the LSD2/NPAC Multimeric Complex.
Cell Rep, 27, 2019
|
|
2Z69
 
 | |
6SEM
 
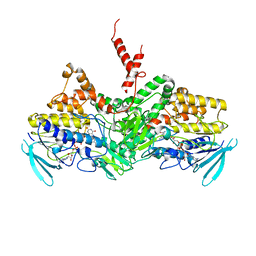 | | Crystal Structure of Ancestral Flavin-containing monooxygenase (FMO) 2 | | Descriptor: | Ancestral Flavin-containing monooxygenase (FMO) 2, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Nicoll, C, Bailleul, G, Fiorentini, F, Mascotti, M.L, Fraaije, M, Mattevi, A. | | Deposit date: | 2019-07-30 | | Release date: | 2019-12-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Ancestral-sequence reconstruction unveils the structural basis of function in mammalian FMOs.
Nat.Struct.Mol.Biol., 27, 2020
|
|
2YG7
 
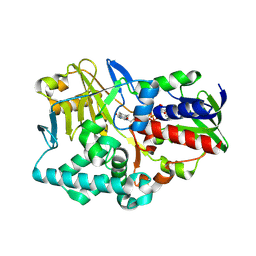 | | Structure-based redesign of cofactor binding in Putrescine Oxidase: A394C-A396T-Q431G Triple mutant | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PUTRESCINE OXIDASE | | Authors: | Kopacz, M.M, Rovida, S, van Duijn, E, Fraaije, M.W, Mattevi, A. | | Deposit date: | 2011-04-11 | | Release date: | 2011-05-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure-Based Redesign of Cofactor Binding in Putrescine Oxidase.
Biochemistry, 50, 2011
|
|
2YG3
 
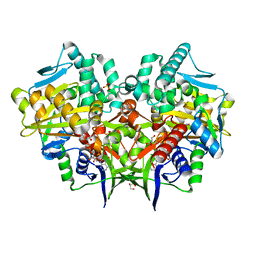 | | Structure-based redesign of cofactor binding in Putrescine Oxidase: wild type enzyme | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, PUTRESCINE OXIDASE, ... | | Authors: | Kopacz, M.M, Rovida, S, van Duijn, E, Fraaije, M.W, Mattevi, A. | | Deposit date: | 2011-04-11 | | Release date: | 2011-05-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Redesign of Cofactor Binding in Putrescine Oxidase.
Biochemistry, 50, 2011
|
|
2YG4
 
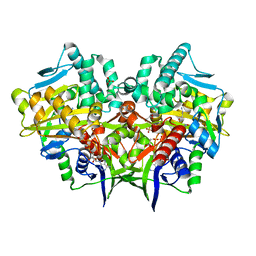 | | Structure-based redesign of cofactor binding in Putrescine Oxidase: wild type bound to Putrescine | | Descriptor: | 4-HYDROXYBUTAN-1-AMINIUM, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Kopacz, M.M, Rovida, S, van Duijn, E, Fraaije, M.W, Mattevi, A. | | Deposit date: | 2011-04-11 | | Release date: | 2011-05-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Redesign of Cofactor Binding in Putrescine Oxidase.
Biochemistry, 50, 2011
|
|
6SF0
 
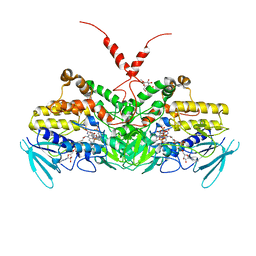 | | Crystal Structure of Ancestral Flavin-containing monooxygenase (FMO) 2 in the presence of NADP+ | | Descriptor: | Ancestral Flavin-containing monooxygenase (FMO) 2, CYCLOHEXYL-HEXYL-BETA-D-MALTOSIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Nicoll, C, Bailleul, G, Fiorentini, F, Mascotti, M.L, Fraaije, M, Mattevi, A. | | Deposit date: | 2019-07-30 | | Release date: | 2019-12-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Ancestral-sequence reconstruction unveils the structural basis of function in mammalian FMOs.
Nat.Struct.Mol.Biol., 27, 2020
|
|
2YG6
 
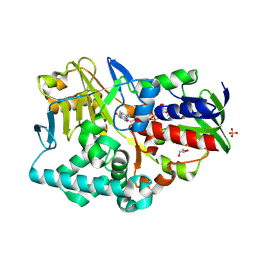 | | Structure-based redesign of cofactor binding in Putrescine Oxidase: P15I-A394C double mutant | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, PUTRESCINE OXIDASE, ... | | Authors: | Kopacz, M.M, Rovida, S, van Duijn, E, Fraaije, M.W, Mattevi, A. | | Deposit date: | 2011-04-11 | | Release date: | 2011-05-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-based redesign of cofactor binding in putrescine oxidase.
Biochemistry, 50, 2011
|
|
6SEK
 
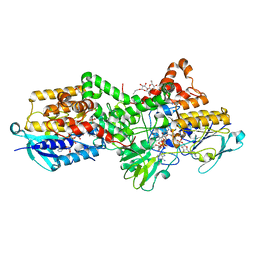 | | Crystal Structure of Ancestral Flavin-containing monooxygenase (FMO) 5 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Ancestral Flavin-containing monooxygenase 5, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Nicoll, C, Bailleul, G, Fiorentini, F, Mascotti, M.L, Fraaije, M, Mattevi, A. | | Deposit date: | 2019-07-30 | | Release date: | 2019-12-25 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Ancestral-sequence reconstruction unveils the structural basis of function in mammalian FMOs.
Nat.Struct.Mol.Biol., 27, 2020
|
|
2YG5
 
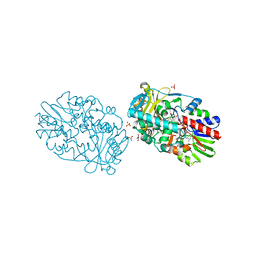 | | Structure-based redesign of cofactor binding in Putrescine Oxidase: A394C mutant | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, PUTRESCINE OXIDASE, ... | | Authors: | Kopacz, M.M, Rovida, S, van Duijn, E, Fraaije, M.W, Mattevi, A. | | Deposit date: | 2011-04-11 | | Release date: | 2011-05-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Based Redesign of Cofactor Binding in Putrescine Oxidase.
Biochemistry, 50, 2011
|
|
6SE3
 
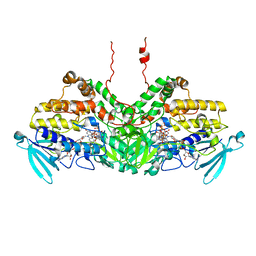 | | Crystal Structure of Ancestral Flavin-containing monooxygenase (FMO) 3-6 | | Descriptor: | Ancestral Flavin-containing monooxygenase (FMO) 3-6, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Nicoll, C, Bailleul, G, Fiorentini, F, Mascotti, M.L, Fraaije, M, Mattevi, A. | | Deposit date: | 2019-07-29 | | Release date: | 2019-12-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Ancestral-sequence reconstruction unveils the structural basis of function in mammalian FMOs.
Nat.Struct.Mol.Biol., 27, 2020
|
|
2NTO
 
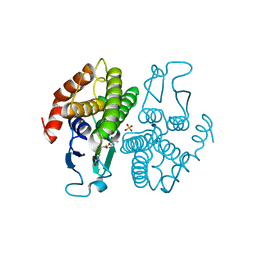 | | Structure of the Glutathione Transferase from Ochrobactrum anthropi in complex with glutathione | | Descriptor: | GLUTATHIONE, SULFATE ION, glutathione S-transferase | | Authors: | Federici, L, Bonivento, D, Di Matteo, A, Allocati, N. | | Deposit date: | 2006-11-08 | | Release date: | 2007-09-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.095 Å) | | Cite: | Role of Ser11 in the stabilization of the structure of Ochrobactrum anthropi glutathione transferase
Biochem.J., 403, 2007
|
|
3GUR
 
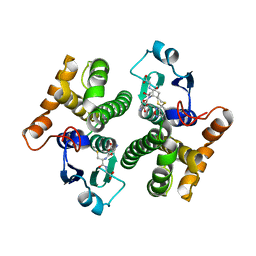 | | Crystal Structure of mu class glutathione S-transferase (GSTM2-2) in complex with glutathione and 6-(7-Nitro-2,1,3-benzoxadiazol-4-ylthio)hexanol (NBDHEX) | | Descriptor: | 1,2-ETHANEDIOL, GLUTATHIONE, Glutathione S-transferase Mu 2, ... | | Authors: | Federici, L, Lo Sterzo, C, Di Matteo, A, Scaloni, F, Federici, G, Caccuri, A.M. | | Deposit date: | 2009-03-30 | | Release date: | 2009-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the binding of the anticancer compound 6-(7-nitro-2,1,3-benzoxadiazol-4-ylthio)hexanol to human glutathione s-transferases
Cancer Res., 69, 2009
|
|
3GUS
 
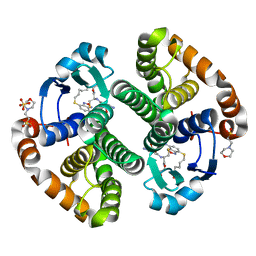 | | Crystal strcture of human Pi class glutathione S-transferase GSTP1-1 in complex with 6-(7-Nitro-2,1,3-benzoxadiazol-4-ylthio)hexanol (NBDHEX) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-[(7-nitro-2,1,3-benzoxadiazol-4-yl)sulfanyl]hexan-1-ol, GLUTATHIONE, ... | | Authors: | Federici, L, Lo Sterzo, C, Di Matteo, A, Scaloni, F, Federici, G, Caccuri, A.M. | | Deposit date: | 2009-03-30 | | Release date: | 2009-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structural basis for the binding of the anticancer compound 6-(7-nitro-2,1,3-benzoxadiazol-4-ylthio)hexanol to human glutathione s-transferases
Cancer Res., 69, 2009
|
|
5LXE
 
 | | F420-dependent glucose-6-phosphate dehydrogenase from Rhodococcus jostii RHA1 | | Descriptor: | F420-dependent glucose-6-phosphate dehydrogenase 1, GLYCEROL, SULFATE ION | | Authors: | Nguyen, Q.-T, Trinco, G, Binda, C, Mattevi, A, Fraaije, M.W. | | Deposit date: | 2016-09-20 | | Release date: | 2016-12-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Discovery and characterization of an F420-dependent glucose-6-phosphate dehydrogenase (Rh-FGD1) from Rhodococcus jostii RHA1.
Appl. Microbiol. Biotechnol., 101, 2017
|
|
5M0Z
 
 | |
5MLN
 
 | |
5MXU
 
 | | Structure of the Y503F mutant of vanillyl alcohol oxidase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Vanillyl-alcohol oxidase | | Authors: | Ewing, T.A, Nguyen, Q.-T, Allan, R.C, Gygli, G, Romero, E, Binda, C, Fraaije, M.W, Mattevi, A, van Berkel, W.J.H. | | Deposit date: | 2017-01-24 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Two tyrosine residues, Tyr-108 and Tyr-503, are responsible for the deprotonation of phenolic substrates in vanillyl-alcohol oxidase.
J. Biol. Chem., 292, 2017
|
|
5MXJ
 
 | | Structure of the Y108F mutant of vanillyl alcohol oxidase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Vanillyl-alcohol oxidase | | Authors: | Ewing, T.A, Nguyen, Q.-T, Allan, R.C, Gygli, G, Romero, E, Binda, C, Fraaije, M.W, Mattevi, A, van Berkel, W.J.H. | | Deposit date: | 2017-01-23 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Two tyrosine residues, Tyr-108 and Tyr-503, are responsible for the deprotonation of phenolic substrates in vanillyl-alcohol oxidase.
J. Biol. Chem., 292, 2017
|
|
5O0X
 
 | | Crystal structure of dehydrogenase domain of Cylindrospermum stagnale NADPH-Oxidase 5 (NOX5) | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Magnani, F, Nenci, S, Mattevi, A. | | Deposit date: | 2017-05-17 | | Release date: | 2017-06-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures and atomic model of NADPH oxidase.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5M10
 
 | |
5O0T
 
 | | CRYSTAL STRUCTURE OF TRANS-MEMBRANE DOMAIN OF Cylindrospermum stagnale NADPH-OXIDASE 5 (NOX5) | | Descriptor: | DECANE, DI(HYDROXYETHYL)ETHER, EICOSANE, ... | | Authors: | Magnani, F, Nenci, S, Mattevi, A. | | Deposit date: | 2017-05-17 | | Release date: | 2017-06-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures and atomic model of NADPH oxidase.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
