6A6B
 
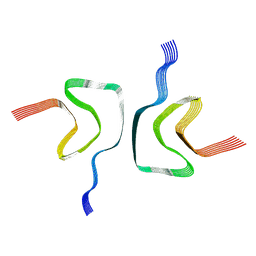 | | cryo-em structure of alpha-synuclein fiber | | Descriptor: | Alpha-synuclein | | Authors: | Li, Y.W, Zhao, C.Y, Luo, F, Liu, Z, Gui, X, Luo, Z, Zhang, X, Li, D, Liu, C, Li, X. | | Deposit date: | 2018-06-27 | | Release date: | 2018-07-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Amyloid fibril structure of alpha-synuclein determined by cryo-electron microscopy
Cell Res., 28, 2018
|
|
8BNZ
 
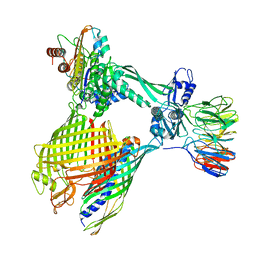 | | BAM-EspP complex structure with BamA-G431C/EspP-N1293C mutations in nanodisc | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | Authors: | Shen, C, Chang, S, Luo, Q, Zhang, Z, Xie, T, Luo, B, Lu, G, Zhu, X, Wei, X, Dong, C, Zhou, R, Zhang, X, Tang, X, Dong, H. | | Deposit date: | 2022-11-14 | | Release date: | 2023-04-26 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of BAM-mediated outer membrane beta-barrel protein assembly.
Nature, 617, 2023
|
|
8BO2
 
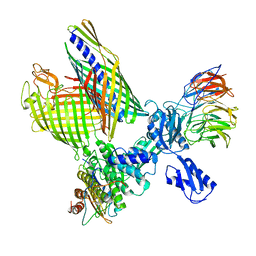 | | BAM-EspP complex structure with BamA-S425C/EspP-S1299C mutations in nanodisc | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | Authors: | Shen, C, Chang, S, Luo, Q, Zhang, Z, Xie, T, Luo, B, Lu, G, Zhu, X, Wei, X, Dong, C, Zhou, R, Zhang, X, Tang, X, Dong, H. | | Deposit date: | 2022-11-14 | | Release date: | 2023-04-26 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of BAM-mediated outer membrane beta-barrel protein assembly.
Nature, 617, 2023
|
|
5ZC3
 
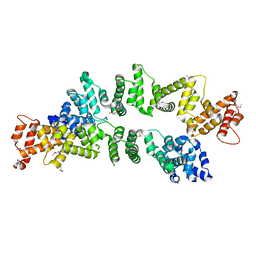 | | The Crystal Structure of PcRxLR12 | | Descriptor: | RxLR effector | | Authors: | Zhao, L, Zhang, X, Zhu, C. | | Deposit date: | 2018-02-14 | | Release date: | 2018-08-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.005 Å) | | Cite: | Crystal structure of the RxLR effector PcRxLR12 from Phytophthora capsici
Biochem. Biophys. Res. Commun., 503, 2018
|
|
6IJJ
 
 | | Photosystem I of Chlamydomonas reinhardtii | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Pan, X, Ma, J, Su, X, Liu, Z, Zhang, X, Li, M. | | Deposit date: | 2018-10-10 | | Release date: | 2019-03-20 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Antenna arrangement and energy transfer pathways of a green algal photosystem-I-LHCI supercomplex.
Nat Plants, 5, 2019
|
|
6IJO
 
 | | Photosystem I of Chlamydomonas reinhardtii | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Pan, X, Ma, J, Su, X, Liu, Z, Zhang, X, Li, M. | | Deposit date: | 2018-10-10 | | Release date: | 2019-03-20 | | Last modified: | 2019-05-01 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Antenna arrangement and energy transfer pathways of a green algal photosystem-I-LHCI supercomplex.
Nat Plants, 5, 2019
|
|
7ARM
 
 | | LolCDE in complex with lipoprotein and LolA | | Descriptor: | (2S)-3-hydroxypropane-1,2-diyl dihexadecanoate, LPP, Lipoprotein-releasing ABC transporter permease subunit LolC, ... | | Authors: | Tang, X.D, Chang, S.H, Zhang, K, Wang, T, Luo, Q.H, Qiao, W, Wang, C, Shen, C.R, Zhang, Z.B, Zhang, Z.Y, Zhu, X.F, Wei, X.W, Dong, C.J, Zhang, X, Dong, H.H. | | Deposit date: | 2020-10-25 | | Release date: | 2021-04-07 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for bacterial lipoprotein relocation by the transporter LolCDE.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7ARL
 
 | | LolCDE in complex with lipoprotein and ADP | | Descriptor: | (2S)-3-hydroxypropane-1,2-diyl dihexadecanoate, ADENOSINE-5'-DIPHOSPHATE, LPP, ... | | Authors: | Tang, X.D, Chang, S.H, Zhang, K, Wang, T, Luo, Q.H, Qiao, W, Wang, C, Shen, C.R, Zhang, Z.B, Zhang, Z.Y, Zhu, X.F, Wei, X.W, Dong, C.J, Zhang, X, Dong, H.H. | | Deposit date: | 2020-10-25 | | Release date: | 2021-04-07 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for bacterial lipoprotein relocation by the transporter LolCDE.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7ARK
 
 | | LolCDE in complex with AMP-PNP in the closed NBD state | | Descriptor: | Lipoprotein-releasing ABC transporter permease subunit LolC, Lipoprotein-releasing system ATP-binding protein LolD, Lipoprotein-releasing system transmembrane protein LolE, ... | | Authors: | Tang, X.D, Chang, S.H, Zhang, K, Wang, T, Luo, Q.H, Qiao, W, Wang, C, Zhang, Z.B, Zhang, Z.Y, Zhu, X.F, Dong, C.J, Zhang, X, Dong, H.H. | | Deposit date: | 2020-10-25 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis for bacterial lipoprotein relocation by the transporter LolCDE.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7ARH
 
 | | LolCDE in complex with lipoprotein | | Descriptor: | (2S)-3-hydroxypropane-1,2-diyl dihexadecanoate, LPP, Lipoprotein-releasing ABC transporter permease subunit LolC, ... | | Authors: | Tang, X.D, Chang, S.H, Zhang, K, Wang, T, Luo, Q.H, Qiao, W, Wang, C, Zhang, Z.B, Zhang, Z.Y, Zhu, X.F, Dong, C.J, Zhang, X, Dong, H.H. | | Deposit date: | 2020-10-25 | | Release date: | 2021-04-07 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for bacterial lipoprotein relocation by the transporter LolCDE.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7ARJ
 
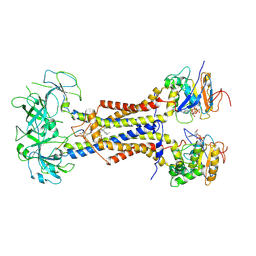 | | LolCDE in complex with lipoprotein and AMPPNP complex undimerized form | | Descriptor: | (2S)-3-hydroxypropane-1,2-diyl dihexadecanoate, LPP, Lipoprotein-releasing ABC transporter permease subunit LolC, ... | | Authors: | Tang, X.D, Chang, S.H, Zhang, K, Wang, T, Luo, Q.H, Qiao, W, Wang, C, Zhang, Z.B, Zhang, Z.Y, Zhu, X.F, Dong, C.J, Zhang, X, Dong, H.H. | | Deposit date: | 2020-10-25 | | Release date: | 2021-04-07 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for bacterial lipoprotein relocation by the transporter LolCDE.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7ARI
 
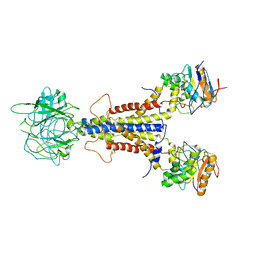 | | LolCDE apo structure | | Descriptor: | Lipoprotein-releasing ABC transporter permease subunit LolC, Lipoprotein-releasing system ATP-binding protein LolD, Lipoprotein-releasing system transmembrane protein LolE | | Authors: | Tang, X.D, Chang, S.H, Zhang, K, Wang, T, Luo, Q.H, Qiao, W, Wang, C, Zhang, Z.B, Zhang, Z.Y, Zhu, X.F, Dong, C.J, Zhang, X, Dong, H.H. | | Deposit date: | 2020-10-25 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for bacterial lipoprotein relocation by the transporter LolCDE.
Nat.Struct.Mol.Biol., 28, 2021
|
|
8R64
 
 | |
1DP4
 
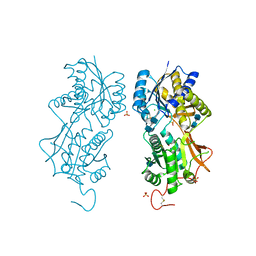 | | DIMERIZED HORMONE BINDING DOMAIN OF THE ATRIAL NATRIURETIC PEPTIDE RECEPTOR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ATRIAL NATRIURETIC PEPTIDE RECEPTOR A, ... | | Authors: | van den Akker, F, Zhang, X, Miyagi, M, Huo, X, Misono, K.S, Yee, V.C. | | Deposit date: | 1999-12-23 | | Release date: | 2000-07-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the dimerized hormone-binding domain of a guanylyl-cyclase-coupled receptor.
Nature, 406, 2000
|
|
4OU1
 
 | | Crystal structure of a computationally designed retro-aldolase covalently bound to folding probe 1 [(6-methoxynaphthalen-2-yl)(oxiran-2-yl)methanol] | | Descriptor: | (1S,2S)-1-(6-methoxynaphthalen-2-yl)propane-1,2-diol, BENZOIC ACID, PHOSPHATE ION, ... | | Authors: | Bhabha, G, Zhang, X, Ekiert, D.C. | | Deposit date: | 2014-02-14 | | Release date: | 2014-03-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Small molecule probes to quantify the functional fraction of a specific protein in a cell with minimal folding equilibrium shifts.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6IPO
 
 | | Ferritin mutant C90A/C102A/C130A/D144C | | Descriptor: | Ferritin heavy chain, MAGNESIUM ION | | Authors: | Zang, J, Chen, H, Zhang, X, Zhao, G. | | Deposit date: | 2018-11-03 | | Release date: | 2019-03-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.998 Å) | | Cite: | Disulfide-mediated conversion of 8-mer bowl-like protein architecture into three different nanocages.
Nat Commun, 10, 2019
|
|
8FNF
 
 | | Cryo-EM structure of RNase-untreated RESC-C in trypanosomal RNA editing | | Descriptor: | Mitochondrial RNA binding complex 1 subunit, Mitochondrial RNA binding protein, Phytanoyl-CoA dioxygenase family protein, ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-27 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
8FNI
 
 | | Cryo-EM structure of RNase-treated RESC-B in trypanosomal RNA editing | | Descriptor: | RNA-editing substrate-binding complex protein 10 (RESC10), RNA-editing substrate-binding complex protein 11 (RESC11), RNA-editing substrate-binding complex protein 13 (RESC13), ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-27 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
8FNK
 
 | | Cryo-EM structure of RNase-untreated RESC-B in trypanosomal RNA editing | | Descriptor: | RNA-editing substrate-binding complex protein 10 (RESC10), RNA-editing substrate-binding complex protein 11 (RESC11), RNA-editing substrate-binding complex protein 13 (RESC13), ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-27 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
8FNC
 
 | | Cryo-EM structure of RNase-treated RESC-C in trypanosomal RNA editing | | Descriptor: | Mitochondrial RNA binding complex 1 subunit, Mitochondrial RNA binding protein, Phytanoyl-CoA dioxygenase family protein, ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-27 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
8FN4
 
 | | Cryo-EM structure of RNase-treated RESC-A in trypanosomal RNA editing | | Descriptor: | RNA-editing substrate-binding complex protein 1 (RESC1), RNA-editing substrate-binding complex protein 2 (RESC2), RNA-editing substrate-binding complex protein 3 (RESC3), ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-26 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
8FN6
 
 | | Cryo-EM structure of RNase-untreated RESC-A in trypanosomal RNA editing | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, RNA-editing substrate-binding complex protein 1 (RESC1), RNA-editing substrate-binding complex protein 2 (RESC2), ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-27 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
6J7G
 
 | |
2LUH
 
 | | NMR structure of the Vta1-Vps60 complex | | Descriptor: | Vacuolar protein sorting-associated protein VTA1, Vacuolar protein-sorting-associated protein 60 | | Authors: | Yang, Z, Vild, C, Ju, J, Zhang, X, Liu, J, Shen, J, Zhao, B, Lan, W, Gong, F, Liu, M, Cao, C, Xu, Z. | | Deposit date: | 2012-06-13 | | Release date: | 2012-11-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of Molecular Recognition between ESCRT-III-like Protein Vps60 and AAA-ATPase Regulator Vta1 in the Multivesicular Body Pathway.
J.Biol.Chem., 287, 2012
|
|
2PZG
 
 | | Minimal human CFTR first nucleotide binding domain as a monomer | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, GLYCEROL, ... | | Authors: | Atwell, S, Conners, K, Emtage, S, Gheyi, T, Glenn, N.R, Hendle, J, Lewis, H.A, Lu, F, Rodgers, L.A, Romero, R, Sauder, J.M, Smith, D, Tien, H, Wasserman, S.R, Zhao, X. | | Deposit date: | 2007-05-18 | | Release date: | 2007-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of a minimal human CFTR first nucleotide-binding domain as a monomer, head-to-tail homodimer, and pathogenic mutant.
Protein Eng.Des.Sel., 23, 2010
|
|
