7YMJ
 
 | | Cryo-EM structure of alpha1AAR-Nb6 complex bound to tamsulosin | | Descriptor: | Nb6, Tamsulosin, alpha1A-adrenergic receptor | | Authors: | Toyoda, Y, Zhu, A, Yan, C, Kobilka, B.K, Liu, X. | | Deposit date: | 2022-07-28 | | Release date: | 2023-07-05 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural basis of alpha 1A -adrenergic receptor activation and recognition by an extracellular nanobody.
Nat Commun, 14, 2023
|
|
7YMH
 
 | | Cryo-EM structure of Nb29-alpha1AAR-miniGsq complex bound to noradrenaline | | Descriptor: | Nb29, Noradrenaline, alpha1A-adrenergic receptor, ... | | Authors: | Toyoda, Y, Zhu, A, Yan, C, Kobilka, B.K, Liu, X. | | Deposit date: | 2022-07-28 | | Release date: | 2023-07-05 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Structural basis of alpha 1A -adrenergic receptor activation and recognition by an extracellular nanobody.
Nat Commun, 14, 2023
|
|
7YM8
 
 | | Cryo-EM structure of Nb29-alpha1AAR-miniGsq complex bound to oxymetazoline | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Oxymetazoline, alpha1A adrenergic receptor, ... | | Authors: | Toyoda, Y, Zhu, A, Yan, C, Kobilka, B.K, Liu, X. | | Deposit date: | 2022-07-27 | | Release date: | 2023-07-05 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Structural basis of alpha 1A -adrenergic receptor activation and recognition by an extracellular nanobody.
Nat Commun, 14, 2023
|
|
7YUF
 
 | | apo human SPNS2 | | Descriptor: | NbFab H-chain, NbFab L-chain, Sphingosine-1-phosphate transporter SPNS2, ... | | Authors: | He, Y, Duan, Y. | | Deposit date: | 2022-08-17 | | Release date: | 2023-09-06 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structural basis of Sphingosine-1-phosphate transport via human SPNS2.
Cell Res., 34, 2024
|
|
7YUD
 
 | | FTY720p-bound human SPNS2 | | Descriptor: | (2~{S})-2-azanyl-4-(4-octylphenyl)-2-[[oxidanyl-bis(oxidanylidene)-$l^{6}-phosphanyl]oxymethyl]butan-1-ol, NbFab L-chain, NbFab-H-chain, ... | | Authors: | He, Y, Duan, Y. | | Deposit date: | 2022-08-17 | | Release date: | 2023-09-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural basis of Sphingosine-1-phosphate transport via human SPNS2.
Cell Res., 34, 2024
|
|
7YUB
 
 | | S1P-bound human SPNS2 | | Descriptor: | (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, NbFab-H-chain, NbFab-L-chain, ... | | Authors: | He, Y, Duan, Y. | | Deposit date: | 2022-08-17 | | Release date: | 2023-09-06 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Structural basis of Sphingosine-1-phosphate transport via human SPNS2.
Cell Res., 34, 2024
|
|
8KAE
 
 | | 16d-bound human SPNS2 | | Descriptor: | 3-[3-(4-decylphenyl)-1,2,4-oxadiazol-5-yl]propan-1-amine, NbFab chain L, NbFab-H chain, ... | | Authors: | He, Y, Duan, Y. | | Deposit date: | 2023-08-03 | | Release date: | 2024-01-03 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structural basis of Sphingosine-1-phosphate transport via human SPNS2.
Cell Res., 34, 2024
|
|
7XV3
 
 | | Cryo-EM structure of LPS-bound GPR174 in complex with Gs protein | | Descriptor: | (2~{S})-2-$l^{4}-azanyl-3-[[(2~{R})-3-octadecanoyloxy-2-oxidanyl-propoxy]-oxidanyl-oxidanylidene-$l^{6}-phosphanyl]oxy-propanoic acid, Engineered G protein subunit S (mini-Gs), Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | He, Y, Liang, J. | | Deposit date: | 2022-05-20 | | Release date: | 2023-02-15 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structural basis of lysophosphatidylserine receptor GPR174 ligand recognition and activation.
Nat Commun, 14, 2023
|
|
8IFO
 
 | | Crystal structure of estrogen related receptor-gamma DNA binding domain complexed with Pla2g12b promoter | | Descriptor: | DNA (5'-D(*GP*AP*GP*GP*AP*CP*AP*AP*AP*GP*GP*TP*GP*AP*AP*AP*C)-3'), DNA (5'-D(*GP*TP*TP*TP*CP*AP*CP*CP*TP*TP*TP*GP*TP*CP*CP*TP*C)-3'), Estrogen-related receptor gamma, ... | | Authors: | Xu, T, Zhen, X, Liu, J. | | Deposit date: | 2023-02-19 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | ERR gamma-DBD undergoes dimerization and conformational rearrangement upon binding to the downstream site of the DR1 element.
Biochem.Biophys.Res.Commun., 656, 2023
|
|
8IYS
 
 | | TUG891-bound FFAR4 in complex with Gq | | Descriptor: | 3-{4-[(4-fluoro-4'-methyl[1,1'-biphenyl]-2-yl)methoxy]phenyl}propanoic acid, Free fatty acid receptor 4, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | He, Y, Yin, H. | | Deposit date: | 2023-04-06 | | Release date: | 2023-06-21 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structural basis of omega-3 fatty acid receptor FFAR4 activation and G protein coupling selectivity.
Cell Res., 33, 2023
|
|
1D1Z
 
 | | CRYSTAL STRUCTURE OF THE XLP PROTEIN SAP | | Descriptor: | SAP SH2 DOMAIN, SULFATE ION | | Authors: | Poy, F, Yaffe, M.B, Sayos, J, Saxena, K, Eck, M.J. | | Deposit date: | 1999-09-22 | | Release date: | 1999-10-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structures of the XLP protein SAP reveal a class of SH2 domains with extended, phosphotyrosine-independent sequence recognition.
Mol.Cell, 4, 1999
|
|
1D4T
 
 | | CRYSTAL STRUCTURE OF THE XLP PROTEIN SAP IN COMPLEX WITH A SLAM PEPTIDE | | Descriptor: | SIGNALING LYMPHOCYTIC ACTIVATION MOLECULE, T CELL SIGNAL TRANSDUCTION MOLECULE SAP | | Authors: | Poy, F, Yaffe, M.B, Sayos, J, Saxena, K, Eck, M.J. | | Deposit date: | 1999-10-06 | | Release date: | 1999-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal structures of the XLP protein SAP reveal a class of SH2 domains with extended, phosphotyrosine-independent sequence recognition.
Mol.Cell, 4, 1999
|
|
6J8F
 
 | | Crystal structure of SVBP-VASH1 with peptide mimic the C-terminal of alpha-tubulin | | Descriptor: | 8-mer peptide, Small vasohibin-binding protein, Tubulinyl-Tyr carboxypeptidase 1 | | Authors: | Liao, S, Gao, J, Xu, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-01-18 | | Release date: | 2019-06-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.283 Å) | | Cite: | Molecular basis of vasohibins-mediated detyrosination and its impact on spindle function and mitosis.
Cell Res., 29, 2019
|
|
6J8N
 
 | | Crystal structure of SVBP-VASH1 complex, mutation C169A of VASH1 | | Descriptor: | Small vasohibin-binding protein, Tubulinyl-Tyr carboxypeptidase 1 | | Authors: | Liao, S, Gao, J, Xu, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-01-20 | | Release date: | 2019-06-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular basis of vasohibins-mediated detyrosination and its impact on spindle function and mitosis.
Cell Res., 29, 2019
|
|
6J9H
 
 | |
6JOM
 
 | | Crystal structure of lipoate protein ligase from Mycoplasma hyopneumoniae | | Descriptor: | 5'-O-[(R)-({5-[(3R)-1,2-DITHIOLAN-3-YL]PENTANOYL}OXY)(HYDROXY)PHOSPHORYL]ADENOSINE, Lipoate--protein ligase | | Authors: | Zhang, H, Chen, H, Ma, G. | | Deposit date: | 2019-03-22 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Functional Identification and Structural Analysis of a New Lipoate Protein Ligase inMycoplasma hyopneumoniae.
Front Cell Infect Microbiol, 10, 2020
|
|
6J91
 
 | | Structure of a hypothetical protease | | Descriptor: | Small vasohibin-binding protein, Tubulinyl-Tyr carboxypeptidase 1 | | Authors: | Liao, S, Gao, J, Xu, C. | | Deposit date: | 2019-01-21 | | Release date: | 2019-06-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Molecular basis of vasohibins-mediated detyrosination and its impact on spindle function and mitosis.
Cell Res., 29, 2019
|
|
2MKX
 
 | |
8Z11
 
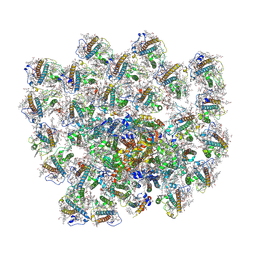 | | Cryo-EM structure of haptophyte photosystem I | | Descriptor: | (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | He, F.Y, Zhao, L.S, Li, K, Zhang, Y.Z, Liu, L.N. | | Deposit date: | 2024-04-10 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Structural insights into the assembly and energy transfer of haptophyte photosystem I-light-harvesting supercomplex.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8ZLZ
 
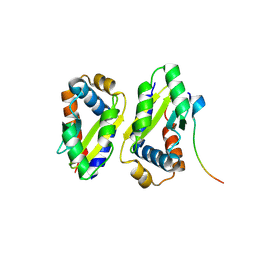 | |
8ZLH
 
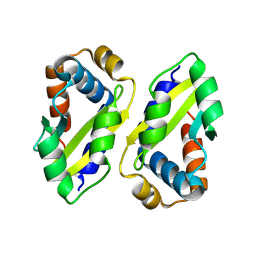 | | The crystal structure of CcmS. | | Descriptor: | All1292 protein | | Authors: | Cheng, J, Li, C.L. | | Deposit date: | 2024-05-20 | | Release date: | 2024-08-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Molecular interactions of the chaperone CcmS and carboxysome shell protein CcmK1 that mediate beta-carboxysome assembly.
Plant Physiol., 196, 2024
|
|
8ZX5
 
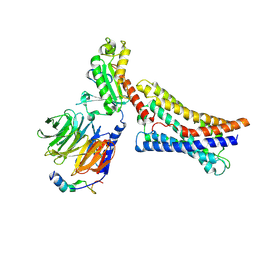 | | AM251-bound GPR55 in complex with G13 | | Descriptor: | 1-(2,4-dichlorophenyl)-5-(4-iodophenyl)-4-methyl-~{N}-piperidin-1-yl-pyrazole-3-carboxamide, G-protein coupled receptor 55, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | He, Y, Xia, R. | | Deposit date: | 2024-06-13 | | Release date: | 2024-11-13 | | Last modified: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structural insight into GPR55 ligand recognition and G-protein coupling.
Cell Res., 35, 2025
|
|
8ZX4
 
 | | LPI-bound GPR55 in complex with G13 | | Descriptor: | G-protein coupled receptor 55, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | He, Y, Xia, R. | | Deposit date: | 2024-06-13 | | Release date: | 2024-11-13 | | Last modified: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural insight into GPR55 ligand recognition and G-protein coupling.
Cell Res., 35, 2025
|
|
7W5M
 
 | |
7WYB
 
 | | ADGRL3/Gi complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | He, Y, Qian, Y. | | Deposit date: | 2022-02-15 | | Release date: | 2022-10-26 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structural insights into adhesion GPCR ADGRL3 activation and G q , G s , G i , and G 12 coupling.
Mol.Cell, 82, 2022
|
|
