7BXH
 
 | | MavC-Lpg2149 complex | | Descriptor: | Lpg2149, MavC | | Authors: | Gao, P, Wang, Y. | | Deposit date: | 2020-04-19 | | Release date: | 2020-05-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Insights into catalysis and regulation of non-canonical ubiquitination and deubiquitination by bacterial deamidase effectors.
Nat Commun, 11, 2020
|
|
7BXF
 
 | | MvcA-Lpg2149 complex | | Descriptor: | Lpg2149, MvcA, PRASEODYMIUM ION | | Authors: | Gao, P. | | Deposit date: | 2020-04-19 | | Release date: | 2020-05-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Insights into catalysis and regulation of non-canonical ubiquitination and deubiquitination by bacterial deamidase effectors.
Nat Commun, 11, 2020
|
|
7BXG
 
 | | MavC-UBE2N-Ub complex | | Descriptor: | MavC, Polyubiquitin-C, Ubiquitin-conjugating enzyme E2 N | | Authors: | Gao, P, Wang, Y. | | Deposit date: | 2020-04-19 | | Release date: | 2020-05-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.705 Å) | | Cite: | Insights into catalysis and regulation of non-canonical ubiquitination and deubiquitination by bacterial deamidase effectors.
Nat Commun, 11, 2020
|
|
3GZN
 
 | | Structure of NEDD8-activating enzyme in complex with NEDD8 and MLN4924 | | Descriptor: | NEDD8, NEDD8-activating enzyme E1 catalytic subunit, NEDD8-activating enzyme E1 regulatory subunit, ... | | Authors: | Sintchak, M.D. | | Deposit date: | 2009-04-07 | | Release date: | 2010-02-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Substrate-assisted inhibition of ubiquitin-like protein-activating enzymes: the NEDD8 E1 inhibitor MLN4924 forms a NEDD8-AMP mimetic in situ.
Mol.Cell, 37, 2010
|
|
8IL0
 
 | | Crystal structure of LmbT from Streptomyces lincolnensis NRRL ISP-5355 | | Descriptor: | Glycosyltransferase | | Authors: | Dai, Y, Li, P, Qiao, H, Xia, M, Liu, W, Fang, P. | | Deposit date: | 2023-03-01 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structural Basis of Low-Molecular-Weight Thiol Glycosylation in Lincomycin A Biosynthesis.
Acs Chem.Biol., 18, 2023
|
|
8SBK
 
 | | Structure of HLA-A*24:02 in complex with peptide, LYLPVRVLI (ATG2A). | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, LEU-TYR-LEU-PRO-VAL-ARG-VAL-LEU-ILE, ... | | Authors: | Mallik, L, Young, M.C, Sgourakis, N.G. | | Deposit date: | 2023-04-03 | | Release date: | 2023-12-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural principles of peptide-centric chimeric antigen receptor recognition guide therapeutic expansion.
Sci Immunol, 8, 2023
|
|
8SBL
 
 | | Structure of HLA-A*24:02 in complex with peptide, LYLPVRVLI | | Descriptor: | Beta-2-microglobulin, LEU-TYR-LEU-PRO-VAL-ARG-VAL-LEU-ILE, MHC class I antigen | | Authors: | Mallik, L, Young, M.C, Sgourakis, N.G. | | Deposit date: | 2023-04-03 | | Release date: | 2023-12-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural principles of peptide-centric chimeric antigen receptor recognition guide therapeutic expansion.
Sci Immunol, 8, 2023
|
|
8SOZ
 
 | | Structure of the complex formed by human interleukin-2 and scFv 602 | | Descriptor: | 602 single chain fragment variable, Interleukin-2 | | Authors: | Gould, J.R, Leonard, E.K, Cao, S.D, Spangler, J.B. | | Deposit date: | 2023-04-30 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Engineered cytokine/antibody fusion proteins improve IL-2 delivery to pro-inflammatory cells and promote antitumor activity.
JCI Insight, 9, 2024
|
|
8SOW
 
 | | Structure of the complex formed by human interleukin-2 and scFv F10 | | Descriptor: | F10 single chain fragment variable, Interleukin-2 | | Authors: | Gould, J.R, Leonard, E.K, Cao, S.D, Spangler, J.B. | | Deposit date: | 2023-04-30 | | Release date: | 2024-09-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Engineered cytokine/antibody fusion proteins improve IL-2 delivery to pro-inflammatory cells and promote antitumor activity.
JCI Insight, 9, 2024
|
|
3PJA
 
 | | Crystal structure of human C3PO complex | | Descriptor: | Translin, Translin-associated protein X | | Authors: | Huang, N, Zhang, H. | | Deposit date: | 2010-11-09 | | Release date: | 2011-05-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of C3PO and mechanism of human RISC activation.
Nat.Struct.Mol.Biol., 18, 2011
|
|
8TVN
 
 | | IRAK4 in complex with compound 23 | | Descriptor: | Interleukin-1 receptor-associated kinase 4, N-{1-[(1R,2S)-2-fluorocyclopropyl]-2-oxo-1,2-dihydropyridin-3-yl}-2-[(1R,4r)-1-methyl-2-oxabicyclo[2.1.1]hexan-4-yl]-6-[(propan-2-yl)oxy]-2H-pyrazolo[3,4-b]pyridine-5-carboxamide | | Authors: | Metrick, C.M, Chodaparambil, J.V. | | Deposit date: | 2023-08-18 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Tiny Pocket Packs a Punch: Leveraging Pyridones for the Discovery of CNS-Penetrant Aza-indazole IRAK4 Inhibitors.
Acs Med.Chem.Lett., 15, 2024
|
|
8TVM
 
 | | IRAK4 in complex with compound 24 | | Descriptor: | Interleukin-1 receptor-associated kinase 4, N-{1-[(1R,2R)-2-fluorocyclopropyl]-2-oxo-1,2-dihydropyridin-3-yl}-2-[(1R,4r)-1-methyl-2-oxabicyclo[2.1.1]hexan-4-yl]-6-[(propan-2-yl)oxy]-2H-pyrazolo[3,4-b]pyridine-5-carboxamide | | Authors: | Metrick, C.M, Chodaparambil, J.V. | | Deposit date: | 2023-08-18 | | Release date: | 2024-06-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Tiny Pocket Packs a Punch: Leveraging Pyridones for the Discovery of CNS-Penetrant Aza-indazole IRAK4 Inhibitors.
Acs Med.Chem.Lett., 15, 2024
|
|
8TYP
 
 | | Complement Protease C1s Inhibited by 6-(4-phenylpiperazin-1-yl)pyridine-3-carboximidamide | | Descriptor: | 6-(4-phenylpiperazin-1-yl)pyridine-3-carboximidamide, Complement C1s subcomponent, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Geisbrecht, B.V. | | Deposit date: | 2023-08-25 | | Release date: | 2023-11-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Inhibition of the C1s Protease and the Classical Complement Pathway by 6-(4-Phenylpiperazin-1-yl)Pyridine-3-Carboximidamide and Chemical Analogs.
J Immunol., 212, 2024
|
|
4OQW
 
 | | Crystal structure of mCardinal far-red fluorescent protein | | Descriptor: | Fluorescent protein FP480 | | Authors: | Burg, J.S, Chu, J, Lam, A.J, Lin, M.Z, Garcia, K.C. | | Deposit date: | 2014-02-10 | | Release date: | 2014-03-12 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Non-invasive intravital imaging of cellular differentiation with a bright red-excitable fluorescent protein.
Nat.Methods, 11, 2014
|
|
4OJ0
 
 | | mCardinal V218E | | Descriptor: | Fluorescent protein FP480 | | Authors: | Ataie, N, Ng, H. | | Deposit date: | 2014-01-20 | | Release date: | 2014-03-19 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Non-invasive intravital imaging of cellular differentiation with a bright red-excitable fluorescent protein.
Nat.Methods, 11, 2014
|
|
3QB5
 
 | | Human C3PO complex in the presence of MnSO4 | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Ye, X, Huang, N, Liu, Y, Paroo, Z, Chen, S, Zhang, H, Liu, Q. | | Deposit date: | 2011-01-12 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of C3PO and mechanism of human RISC activation.
Nat.Struct.Mol.Biol., 18, 2011
|
|
4F2L
 
 | | Structure of a regulatory domain of AMPK | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, MAGNESIUM ION | | Authors: | Xin, F.J, Zhang, Y.Y, Wang, J, Wang, Z.X, Wu, J.W. | | Deposit date: | 2012-05-08 | | Release date: | 2013-06-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Conserved regulatory elements in AMPK
Nature, 498, 2013
|
|
5W2E
 
 | | HCV NS5B RNA-dependent RNA polymerase in complex with non-nucleoside inhibitor MK-8876 | | Descriptor: | 2-(4-fluorophenyl)-5-(11-fluoro-6H-pyrido[2',3':5,6][1,3]oxazino[3,4-a]indol-2-yl)-N-methyl-6-[methyl(methylsulfonyl)amino]-1-benzofuran-3-carboxamide, Genome polyprotein | | Authors: | Lesburg, C.A, Ummat, A. | | Deposit date: | 2017-06-06 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Development of a New Structural Class of Broadly Acting HCV Non-Nucleoside Inhibitors Leading to the Discovery of MK-8876.
ChemMedChem, 12, 2017
|
|
5W4S
 
 | |
4P8O
 
 | | S. aureus gyrase bound to an aminobenzimidazole urea inhibitor | | Descriptor: | 1-ethyl-3-[5-(5-fluoropyridin-3-yl)-7-(pyrimidin-2-yl)-1H-benzimidazol-2-yl]urea, DNA gyrase subunit B | | Authors: | Jacobs, M.D. | | Deposit date: | 2014-03-31 | | Release date: | 2014-10-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Second-generation antibacterial benzimidazole ureas: discovery of a preclinical candidate with reduced metabolic liability.
J.Med.Chem., 57, 2014
|
|
4QR8
 
 | | Crystal Structure of E coli pepQ | | Descriptor: | MAGNESIUM ION, Xaa-Pro dipeptidase | | Authors: | Pingwei, L. | | Deposit date: | 2014-06-30 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Structural basis of substrate selectivity of E. coli prolidase.
Plos One, 9, 2014
|
|
8TX0
 
 | | IRAK4 in complex with compound | | Descriptor: | 1,2-ETHANEDIOL, Interleukin-1 receptor-associated kinase 4, SODIUM ION, ... | | Authors: | Metrick, C.M, Chodaparambil, J.V. | | Deposit date: | 2023-08-21 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of BIO-8169─A Highly Potent, Selective, and Brain-Penetrant IRAK4 Inhibitor for the Treatment of Neuroinflammation.
J.Med.Chem., 67, 2024
|
|
8U0H
 
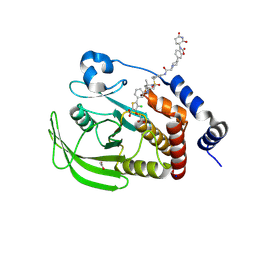 | | Crystal structure of PTPN2 with a PROTAC | | Descriptor: | (5P)-3-(carboxymethoxy)-4-chloro-5-(3-{[(4S)-1-({3-[2-(4-{3-[(3R)-2,6-dioxopiperidin-3-yl]-2-oxo-2,3-dihydro-1,3-benzoxazol-6-yl}piperidin-1-yl)acetamido]phenyl}methanesulfonyl)-2,2-dimethylpiperidin-4-yl]amino}phenyl)thiophene-2-carboxylic acid, ACETATE ION, PTPN2, ... | | Authors: | Jain, R, Longenecker, K, Qiu, W. | | Deposit date: | 2023-08-29 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Mechanistic insights into a heterobifunctional degrader-induced PTPN2/N1 complex.
Commun Chem, 7, 2024
|
|
8UH6
 
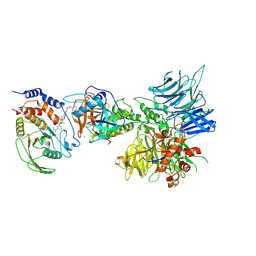 | | Degrader-induced complex between PTPN2 and CRBN-DDB1 | | Descriptor: | (5P)-3-(carboxymethoxy)-4-chloro-5-(3-{[(4S)-1-({3-[(4-{1-[(3R)-2,6-dioxopiperidin-3-yl]-3-methyl-2-oxo-2,3-dihydro-1H-benzimidazol-5-yl}piperidine-1-carbonyl)amino]phenyl}methanesulfonyl)-2,2-dimethylpiperidin-4-yl]amino}phenyl)thiophene-2-carboxylic acid, DNA damage-binding protein 1, Protein cereblon, ... | | Authors: | Catalano, C, Bratkowski, M, Scapin, G, Hao, Q. | | Deposit date: | 2023-10-06 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mechanistic insights into a heterobifunctional degrader-induced PTPN2/N1 complex.
Commun Chem, 7, 2024
|
|
8GS1
 
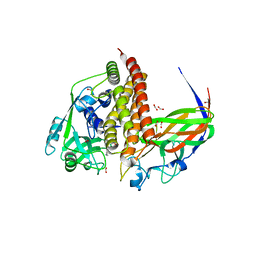 | | Crystal structure of AziU2-U3 complex from Streptomyces sahachiroi NRRL2485 | | Descriptor: | Azi28, Azi29, FORMIC ACID, ... | | Authors: | Cheng, Y, Li, P, Liu, W, Fang, P. | | Deposit date: | 2022-09-04 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Oxidase Heterotetramer Completes 1-Azabicyclo[3.1.0]hexane Formation with the Association of a Nonribosomal Peptide Synthetase.
J.Am.Chem.Soc., 145, 2023
|
|
