3O05
 
 | | Crystal Structure of Yeast Pyridoxal 5-Phosphate Synthase Snz1 Complxed with Substrate PLP | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Pyridoxine biosynthesis protein SNZ1 | | Authors: | Teng, Y.B, Zhang, X, He, Y.X, Hu, H.X, Zhou, C.Z. | | Deposit date: | 2010-07-19 | | Release date: | 2010-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into the catalytic mechanism of the yeast pyridoxal 5-phosphate synthase Snz1
Biochem.J., 432, 2010
|
|
1J04
 
 | | Structural mechanism of enzyme mistargeting in hereditary kidney stone disease in vitro | | Descriptor: | (AMINOOXY)ACETIC ACID, GLYCEROL, alanine--glyoxylate aminotransferase | | Authors: | Zhang, X, Djordjevic, S, Bartlam, M, Ye, S, Rao, Z, Danpure, C.J. | | Deposit date: | 2002-10-30 | | Release date: | 2003-11-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural implications of a G170R mutation of alanine:glyoxylate aminotransferase that is associated with peroxisome-to-mitochondrion mistargeting.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
7YSK
 
 | | Crystal structure of D-Cysteine desulfhydrase from Pectobacterium atrosepticum | | Descriptor: | D-Cysteine desulfhydrase | | Authors: | Zhang, X, Wang, L, Xu, X, Xing, X, Zhou, J. | | Deposit date: | 2022-08-12 | | Release date: | 2022-12-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization and structural basis of D-cysteine desulfhydrase from Pectobacterium atrosepticum
Tetrahedron, 2022
|
|
7YSL
 
 | | Crystal structure of D-Cysteine desulfhydrase with a trapped PLP-pyruvate geminal diamine | | Descriptor: | 1,2-ETHANEDIOL, D-Cysteine desulfhydrase, FORMIC ACID | | Authors: | Zhang, X, Wang, L, Xu, X, Xing, X, Zhou, J. | | Deposit date: | 2022-08-12 | | Release date: | 2022-12-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Characterization and structural basis of D-cysteine desulfhydrase from Pectobacterium atrosepticum
Tetrahedron, 2022
|
|
5GQH
 
 | | Cryo-EM structure of PaeCas3-AcrF3 complex | | Descriptor: | CRISPR-associated nuclease/helicase Cas3 subtype I-F/YPEST, anti-CRISPR protein 3 | | Authors: | Zhang, X, Ma, J, Wang, Y, Wang, J. | | Deposit date: | 2016-08-07 | | Release date: | 2016-09-21 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | A CRISPR evolutionary arms race: structural insights into viral anti-CRISPR/Cas responses
Cell Res., 26, 2016
|
|
2Z41
 
 | | Crystal Structure Analysis of the Ski2-type RNA helicase | | Descriptor: | MAGNESIUM ION, putative ski2-type helicase | | Authors: | Nakashima, T, Zhang, X, Kakuta, Y, Yao, M, Tanaka, I, Kimura, M. | | Deposit date: | 2007-06-12 | | Release date: | 2008-04-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | Crystal structure of an archaeal Ski2p-like protein from Pyrococcus horikoshii OT3
Protein Sci., 17, 2008
|
|
5NWT
 
 | | Crystal Structure of Escherichia coli RNA polymerase - Sigma54 Holoenzyme complex | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zhang, X, Buck, M, Darbari, V.C, Yang, Y, Zhang, N, Lu, D, Glyde, R, Wang, Y, Winkelman, J, Gourse, R.L, Murakami, K.S. | | Deposit date: | 2017-05-08 | | Release date: | 2017-09-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.76 Å) | | Cite: | TRANSCRIPTION. Structures of the RNA polymerase-Sigma54 reveal new and conserved regulatory strategies.
Science, 349, 2015
|
|
8WRQ
 
 | |
1JW2
 
 | | SOLUTION STRUCTURE OF HEMOLYSIN EXPRESSION MODULATING PROTEIN Hha FROM ESCHERICHIA COLI. Ontario Centre for Structural Proteomics target EC0308_1_72; Northeast Structural Genomics Target ET88 | | Descriptor: | HEMOLYSIN EXPRESSION MODULATING PROTEIN Hha | | Authors: | Chang, X, Yee, A, Savchenko, A, Edwards, A.M, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2001-09-02 | | Release date: | 2002-02-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An NMR approach to structural proteomics
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
2QVJ
 
 | | Crystal structure of a vesicular stomatitis virus nucleocapsid protein Ser290Trp mutant | | Descriptor: | Nucleocapsid protein | | Authors: | Luo, M, Green, T.J, Zhang, X, Tsao, J, Qiu, S. | | Deposit date: | 2007-08-08 | | Release date: | 2008-01-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Role of intermolecular interactions of vesicular stomatitis virus nucleoprotein in RNA encapsidation.
J.Virol., 82, 2008
|
|
1JW3
 
 | | Solution Structure of Methanobacterium Thermoautotrophicum Protein 1598. Ontario Centre for Structural Proteomics target MTH1598_1_140; Northeast Structural Genomics Target TT6 | | Descriptor: | Conserved Hypothetical Protein MTH1598 | | Authors: | Chang, X, Connelly, G, Yee, A, Kennedy, M.A, Edwards, A.M, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2001-09-02 | | Release date: | 2002-02-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An NMR approach to structural proteomics.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
3O06
 
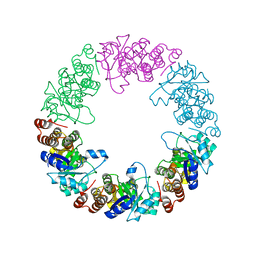 | |
3O07
 
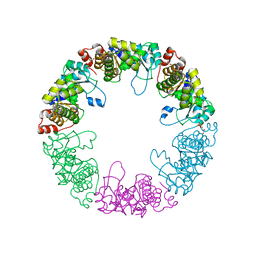 | | Crystal structure of yeast pyridoxal 5-phosphate synthase Snz1 complexed with substrate G3P | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE, Pyridoxine biosynthesis protein SNZ1 | | Authors: | Teng, Y.B, Zhang, X, Hu, H.X, Zhou, C.Z. | | Deposit date: | 2010-07-19 | | Release date: | 2010-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the catalytic mechanism of the yeast pyridoxal 5-phosphate synthase Snz1
Biochem.J., 432, 2010
|
|
8INB
 
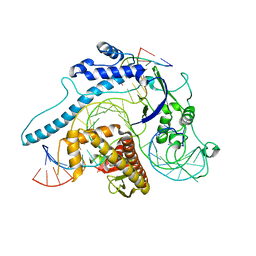 | | Cryo-EM structure of Cas12j-SF05-crRNA-dsDNA complex | | Descriptor: | Cas12j-SF05, NTS-DNA, TS-DNA, ... | | Authors: | Zhang, X, Duan, Z.Q, Zhu, J.K. | | Deposit date: | 2023-03-09 | | Release date: | 2024-03-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular basis for DNA cleavage by the hypercompact Cas12j-SF05.
Cell Discov, 9, 2023
|
|
8Z0O
 
 | | Cryo-EM structure of chitin synthase | | Descriptor: | MANGANESE (II) ION, URIDINE-5'-DIPHOSPHATE, chitin synthase | | Authors: | Zhang, X, Niu, S, Li, P, Bi, Y. | | Deposit date: | 2024-04-10 | | Release date: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Chitin Translocation Is Functionally Coupled with Synthesis in Chitin Synthase.
Int J Mol Sci, 25, 2024
|
|
4RSQ
 
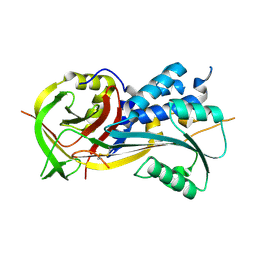 | | 2.9A resolution structure of SRPN2 (K198C/E359C) from Anopheles gambiae | | Descriptor: | Serpin 2 | | Authors: | Lovell, S, Battaile, K.P, Zhang, X, Meekins, D.A, An, C, Michel, K. | | Deposit date: | 2014-11-10 | | Release date: | 2014-12-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and Inhibitory Effects of Hinge Loop Mutagenesis in Serpin-2 from the Malaria Vector Anopheles gambiae.
J.Biol.Chem., 290, 2015
|
|
4ROA
 
 | | 1.90A resolution structure of SRPN2 (S358W) from Anopheles gambiae | | Descriptor: | Serpin 2 | | Authors: | Lovell, S, Battaile, K.P, Zhang, X, Meekins, D.A, An, C, Michel, K. | | Deposit date: | 2014-10-28 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Inhibitory Effects of Hinge Loop Mutagenesis in Serpin-2 from the Malaria Vector Anopheles gambiae.
J.Biol.Chem., 290, 2015
|
|
8AEB
 
 | | SARS-CoV-2 Main Protease complexed with N-(pyridin-3-ylmethyl)thioformamide | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, N-(pyridin-3-ylmethyl)thioformamide, ... | | Authors: | Hanoulle, X, Charton, J, Deprez, B. | | Deposit date: | 2022-07-12 | | Release date: | 2023-03-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Novel dithiocarbamates selectively inhibit 3CL protease of SARS-CoV-2 and other coronaviruses.
Eur.J.Med.Chem., 250, 2023
|
|
1RHP
 
 | |
4RO9
 
 | | 2.0A resolution structure of SRPN2 (S358E) from Anopheles gambiae | | Descriptor: | GLYCEROL, Serpin 2 | | Authors: | Lovell, S, Battaile, K.P, Zhang, X, Meekins, D.A, An, C, Michel, K. | | Deposit date: | 2014-10-28 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Inhibitory Effects of Hinge Loop Mutagenesis in Serpin-2 from the Malaria Vector Anopheles gambiae.
J.Biol.Chem., 290, 2015
|
|
4ROB
 
 | | 2.8A resolution structure of SRPN2 (K198C) from Anopheles gambiae | | Descriptor: | Serpin 2 | | Authors: | Lovell, S, Battaile, K.P, Zhang, X, Meekins, D.A, An, C, Michel, K. | | Deposit date: | 2014-10-28 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | 2.8A resolution structure of SRPN2 (K198C) from Anopheles gambiae
To be Published
|
|
3RYT
 
 | | The Plexin A1 intracellular region in complex with Rac1 | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Plexin-A1, ... | | Authors: | Zhang, X, He, H. | | Deposit date: | 2011-05-11 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.582 Å) | | Cite: | Plexins Are GTPase-Activating Proteins for Rap and Are Activated by Induced Dimerization.
Sci.Signal., 5, 2012
|
|
7LCI
 
 | | PF 06882961 bound to the glucagon-like peptide-1 receptor (GLP-1R):Gs complex | | Descriptor: | 2-[(4-{6-[(4-cyano-2-fluorophenyl)methoxy]pyridin-2-yl}piperidin-1-yl)methyl]-1-{[(2S)-oxetan-2-yl]methyl}-1H-benzimidazole-6-carboxylic acid, Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Belousoff, M.J, Johnson, R.M, Drulyte, I, Yu, L, Kotecha, A, Danev, R, Wootten, D, Zhang, X, Sexton, P.M. | | Deposit date: | 2021-01-11 | | Release date: | 2021-01-20 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Evolving cryo-EM structural approaches for GPCR drug discovery.
Structure, 29, 2021
|
|
5YC0
 
 | | Crystal structure of LP-46/N44 | | Descriptor: | Envelope glycoprotein, LP-46 | | Authors: | Zhang, X, Wang, X, He, Y. | | Deposit date: | 2017-09-05 | | Release date: | 2018-02-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Exceptional potency and structural basis of a T1249-derived lipopeptide fusion inhibitor against HIV-1, HIV-2, and simian immunodeficiency virus
J. Biol. Chem., 293, 2018
|
|
7CTH
 
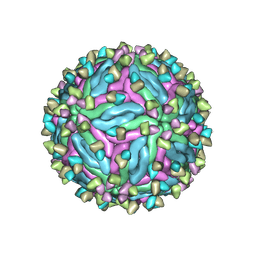 | | Cryo-EM structure of dengue virus serotype 2 in complex with the scFv fragment of the broadly neutralizing antibody EDE1 C10 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Core protein, Single Chain Variable Fragment | | Authors: | Zhang, X, Sharma, A, Duquerroy, S, Zhou, Z.H, Rey, F.A. | | Deposit date: | 2020-08-19 | | Release date: | 2021-12-01 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The epitope arrangement on flavivirus particles contributes to Mab C10's extraordinary neutralization breadth across Zika and dengue viruses.
Cell, 184, 2021
|
|
