6W26
 
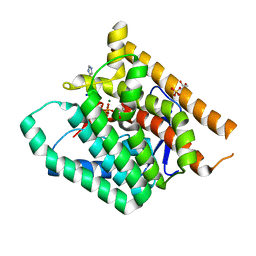 | | Terpenoid Cyclase FgGS in Complex with Mg, Inorganic Pyrophosphate, and Imidazole | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, IMIDAZOLE, ... | | Authors: | Herbst-Gervasoni, C.J, Christianson, D.W. | | Deposit date: | 2020-03-05 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of the cryptic function of terpene cyclases as aromatic prenyltransferases.
Nat Commun, 11, 2020
|
|
6UFO
 
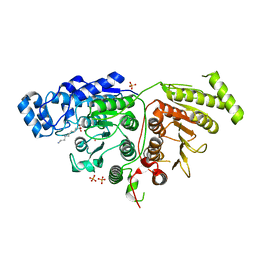 | |
6UO4
 
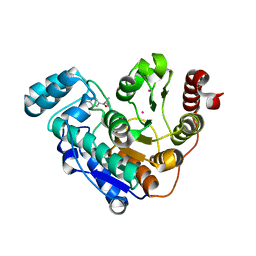 | |
6UIL
 
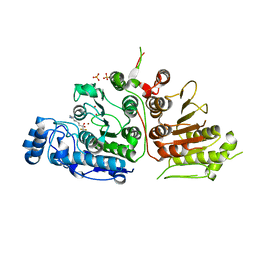 | | Crystal Structure of Danio rerio Histone Deacetylase 10 in Complex with 7-[(3-aminopropyl)amino]-1,1,1-trifluoroheptan-2-one | | Descriptor: | 7-[(3-aminopropyl)amino]-1,1,1-trifluoroheptane-2,2-diol, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Herbst-Gervasoni, C.J, Christianson, D.W. | | Deposit date: | 2019-10-01 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Binding ofN8-Acetylspermidine Analogues to Histone Deacetylase 10 Reveals Molecular Strategies for Blocking Polyamine Deacetylation.
Biochemistry, 58, 2019
|
|
6UOC
 
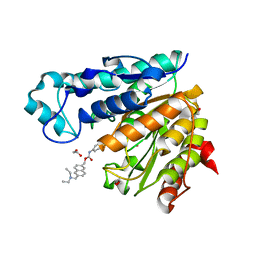 | | Crystal structure of Danio rerio histone deacetylase 6 catalytic domain 1 (CD1) K330L mutant complexed with Givinostat | | Descriptor: | 1,2-ETHANEDIOL, Givinostat, Histone deacetylase 6, ... | | Authors: | Osko, J.D, Christianson, D.W. | | Deposit date: | 2019-10-14 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.400008 Å) | | Cite: | Structural Basis of Catalysis and Inhibition of HDAC6 CD1, the Enigmatic Catalytic Domain of Histone Deacetylase 6.
Biochemistry, 58, 2019
|
|
6UOB
 
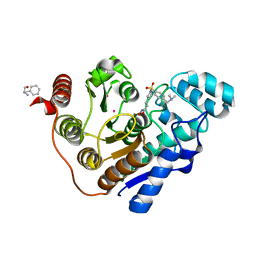 | | Crystal structure of Danio rerio histone deacetylase 6 catalytic domain 1 (CD1) K330L mutant complexed with Resminostat | | Descriptor: | 1,2-ETHANEDIOL, Histone deacetylase 6, PHENYLALANINE, ... | | Authors: | Osko, J.D, Christianson, D.W. | | Deposit date: | 2019-10-14 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.58000576 Å) | | Cite: | Structural Basis of Catalysis and Inhibition of HDAC6 CD1, the Enigmatic Catalytic Domain of Histone Deacetylase 6.
Biochemistry, 58, 2019
|
|
6WBQ
 
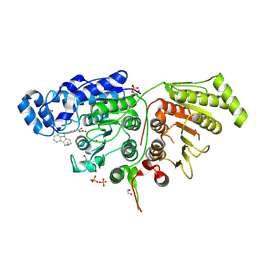 | | Crystal Structure of Danio rerio Histone Deacetylase 10 in Complex with Tubastatin A | | Descriptor: | 1,2-ETHANEDIOL, 4-[(2-methyl-3,4-dihydro-1~{H}-pyrido[4,3-b]indol-5-yl)methyl]-~{N}-oxidanyl-benzamide, PHOSPHATE ION, ... | | Authors: | Herbst-Gervasoni, C.J, Christianson, D.W. | | Deposit date: | 2020-03-27 | | Release date: | 2020-07-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for the Selective Inhibition of HDAC10, the Cytosolic Polyamine Deacetylase.
Acs Chem.Biol., 15, 2020
|
|
6VNR
 
 | | Crystal Structure of Danio rerio Histone Deacetylase 6 Catalytic Domain 2 (CD2) Complexed with Bishydroxamic Acid Inhibitor | | Descriptor: | 1,2-ETHANEDIOL, Hdac6 protein, N-hydroxy-1-{[4-(hydroxycarbamoyl)phenyl]methyl}-1H-indole-6-carboxamide, ... | | Authors: | Osko, J.D, Porter, N.J, Christianson, D.W. | | Deposit date: | 2020-01-29 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.94301319 Å) | | Cite: | Design and Synthesis of Dihydroxamic Acids as HDAC6/8/10 Inhibitors.
Chemmedchem, 15, 2020
|
|
4RN1
 
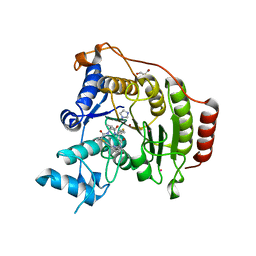 | | Crystal structure of S39D HDAC8 in complex with a largazole analogue. | | Descriptor: | (5R,8S,11S)-5-methyl-8-(propan-2-yl)-11-[(1E)-4-sulfanylbut-1-en-1-yl]-3-thia-7,10,14,20,21-pentaazatricyclo[14.3.1.1~2,5~]henicosa-1(20),2(21),16,18-tetraene-6,9,13-trione, GLYCEROL, Histone deacetylase 8, ... | | Authors: | Decroos, C, Christianson, D.W. | | Deposit date: | 2014-10-22 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Variable Active Site Loop Conformations Accommodate the Binding of Macrocyclic Largazole Analogues to HDAC8.
Biochemistry, 54, 2015
|
|
4RN0
 
 | | Crystal structure of S39D HDAC8 in complex with a largazole analogue. | | Descriptor: | (5R,8S,11S)-5-methyl-8-(propan-2-yl)-11-[(1E)-4-sulfanylbut-1-en-1-yl]-3,17-dithia-7,10,14,19,20-pentaazatricyclo[14.2.1.1~2,5~]icosa-1(18),2(20),16(19)-triene-6,9,13-trione, GLYCEROL, Histone deacetylase 8, ... | | Authors: | Decroos, C, Christianson, D.W. | | Deposit date: | 2014-10-22 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.761 Å) | | Cite: | Variable Active Site Loop Conformations Accommodate the Binding of Macrocyclic Largazole Analogues to HDAC8.
Biochemistry, 54, 2015
|
|
1BNT
 
 | | CARBONIC ANHYDRASE II INHIBITOR | | Descriptor: | 3,4-DIHYDRO-4-HYDROXY-2-(4-METHOXYPHENYL)-2H-THIENO[3,2-E]-1,2-THIAZINE-6-SULFONAMIDE-1,1-DIOXIDE, CARBONIC ANHYDRASE, MERCURY (II) ION, ... | | Authors: | Boriack-Sjodin, P.A, Zeitlin, S, Christianson, D.W. | | Deposit date: | 1998-07-30 | | Release date: | 1999-06-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural analysis of inhibitor binding to human carbonic anhydrase II.
Protein Sci., 7, 1998
|
|
6WDX
 
 | |
6WDV
 
 | |
1BNU
 
 | | CARBONIC ANHYDRASE II INHIBITOR | | Descriptor: | 3,4-DIHYDRO-4-HYDROXY-2-(2-THIENYMETHYL)-2H-THIENO[3,2-E]-1,2-THIAZINE-6-SULFONAMIDE-1,1-DIOXIDE, CARBONIC ANHYDRASE, MERCURY (II) ION, ... | | Authors: | Boriack-Sjodin, P.A, Zeitlin, S, Christianson, D.W. | | Deposit date: | 1998-07-30 | | Release date: | 1999-06-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural analysis of inhibitor binding to human carbonic anhydrase II.
Protein Sci., 7, 1998
|
|
1BNW
 
 | | CARBONIC ANHYDRASE II INHIBITOR | | Descriptor: | CARBONIC ANHYDRASE, MERCURY (II) ION, N-(2-THIENYLMETHYL)-2,5-THIOPHENEDISULFONAMIDE, ... | | Authors: | Boriack-Sjodin, P.A, Zeitlin, S, Christianson, D.W. | | Deposit date: | 1998-07-30 | | Release date: | 1999-06-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural analysis of inhibitor binding to human carbonic anhydrase II.
Protein Sci., 7, 1998
|
|
1BNM
 
 | | CARBONIC ANHYDRASE II INHIBITOR | | Descriptor: | (R)-3,4-DIHYDRO-2-(3-METHOXYPHENYL)-4-METHYLAMINO-2H-THIENO[3,2-E]-1,2-THIAZINE-6-SULFONAMIDE-1,1-DIOXIDE, CARBONIC ANHYDRASE, MERCURY (II) ION, ... | | Authors: | Boriack-Sjodin, P.A, Zeitlin, S, Christianson, D.W. | | Deposit date: | 1998-07-30 | | Release date: | 1999-05-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural analysis of inhibitor binding to human carbonic anhydrase II.
Protein Sci., 7, 1998
|
|
1BN3
 
 | | CARBONIC ANHYDRASE II INHIBITOR | | Descriptor: | 2-(3-METHOXYPHENYL)-2H-THIENO-[3,2-E]-1,2-THIAZINE-6-SULFINAMIDE-1,1-DIOXIDE, CARBONIC ANHYDRASE, MERCURY (II) ION, ... | | Authors: | Boriack-Sjodin, P.A, Zeitlin, S, Christianson, D.W. | | Deposit date: | 1998-07-31 | | Release date: | 1999-05-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural analysis of inhibitor binding to human carbonic anhydrase II.
Protein Sci., 7, 1998
|
|
1BNN
 
 | | CARBONIC ANHYDRASE II INHIBITOR | | Descriptor: | 3,,4-DIHYDRO-2-(3-METHOXYPHENYL)-2H-THIENO-[3,2-E]-1,2-THIAZINE-6-SULFONAMIDE-1,1-DIOXIDE, CARBONIC ANHYDRASE, MERCURY (II) ION, ... | | Authors: | Boriack-Sjodin, P.A, Zeitlin, S, Christianson, D.W. | | Deposit date: | 1998-07-30 | | Release date: | 1999-05-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of inhibitor binding to human carbonic anhydrase II.
Protein Sci., 7, 1998
|
|
1CA3
 
 | |
4Q3P
 
 | | Crystal structure of Schistosoma mansoni arginase | | Descriptor: | Arginase, GLYCEROL, MANGANESE (II) ION | | Authors: | Hai, Y, Edwards, J.E, Van Zandt, M.C, Hoffmann, K.F, Christianson, D.W. | | Deposit date: | 2014-04-12 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Crystal Structure of Schistosoma mansoni Arginase, a Potential Drug Target for the Treatment of Schistosomiasis.
Biochemistry, 53, 2014
|
|
4Q3V
 
 | | Crystal structure of Schistosoma mansoni arginase in complex with inhibitor BEC | | Descriptor: | Arginase, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Hai, Y, Edwards, J.E, Van Zandt, M.C, Hoffmann, K.F, Christianson, D.W. | | Deposit date: | 2014-04-12 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Schistosoma mansoni Arginase, a Potential Drug Target for the Treatment of Schistosomiasis.
Biochemistry, 53, 2014
|
|
4Q41
 
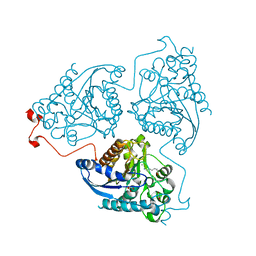 | | Crystal structure of Schistosoma mansoni arginase in complex with L-lysine | | Descriptor: | Arginase, GLYCEROL, LYSINE, ... | | Authors: | Hai, Y, Edwards, J.E, Van Zandt, M.C, Hoffmann, K.F, Christianson, D.W. | | Deposit date: | 2014-04-12 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Crystal Structure of Schistosoma mansoni Arginase, a Potential Drug Target for the Treatment of Schistosomiasis.
Biochemistry, 53, 2014
|
|
4Q3Q
 
 | | Crystal structure of Schistosoma mansoni arginase in complex with inhibitor ABH | | Descriptor: | 2(S)-AMINO-6-BORONOHEXANOIC ACID, Arginase, GLYCEROL, ... | | Authors: | Hai, Y, Edwards, J.E, Van Zandt, M.C, Hoffmann, K.F, Christianson, D.W. | | Deposit date: | 2014-04-12 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Crystal Structure of Schistosoma mansoni Arginase, a Potential Drug Target for the Treatment of Schistosomiasis.
Biochemistry, 53, 2014
|
|
4Q3S
 
 | | Crystal structure of Schistosoma mansoni arginase in complex with inhibitor ABHPE | | Descriptor: | Arginase, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Hai, Y, Edwards, J.E, Van Zandt, M.C, Hoffmann, K.F, Christianson, D.W. | | Deposit date: | 2014-04-12 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal Structure of Schistosoma mansoni Arginase, a Potential Drug Target for the Treatment of Schistosomiasis.
Biochemistry, 53, 2014
|
|
4Q3T
 
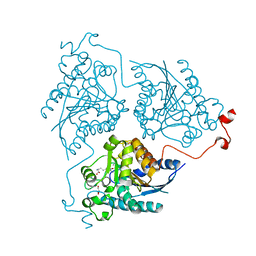 | | Crystal structure of Schistosoma mansoni arginase in complex with inhibitor NOHA | | Descriptor: | Arginase, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Hai, Y, Edwards, J.E, Van Zandt, M.C, Hoffmann, K.F, Christianson, D.W. | | Deposit date: | 2014-04-12 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.142 Å) | | Cite: | Crystal Structure of Schistosoma mansoni Arginase, a Potential Drug Target for the Treatment of Schistosomiasis.
Biochemistry, 53, 2014
|
|
