7W55
 
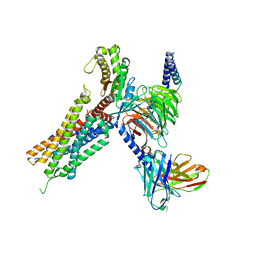 | | Cryo-EM structure of the neuromedin U-bound neuromedin U receptor 2-Gq protein complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(q) subunit alpha, ... | | Authors: | You, C, Xu, H.E, Jiang, Y. | | Deposit date: | 2021-11-29 | | Release date: | 2022-04-20 | | Last modified: | 2022-06-15 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into the peptide selectivity and activation of human neuromedin U receptors.
Nat Commun, 13, 2022
|
|
7W53
 
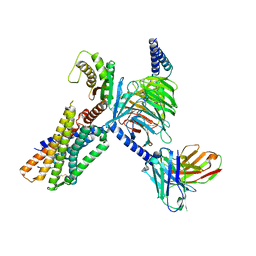 | | Cryo-EM structure of the neuromedin U-bound neuromedin U receptor 1-Gq protein complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(q) subunit alpha, ... | | Authors: | You, C, Xu, H.E, Jiang, Y. | | Deposit date: | 2021-11-29 | | Release date: | 2022-04-20 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into the peptide selectivity and activation of human neuromedin U receptors.
Nat Commun, 13, 2022
|
|
5TUR
 
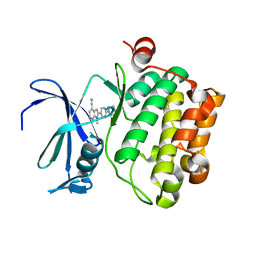 | | Pim-1 kinase in complex with a 7-azaindole | | Descriptor: | 1-methyl-2-[4-(piperazin-1-yl)phenyl]-1H-pyrrolo[2,3-b]pyridine-4-carbonitrile, Serine/threonine-protein kinase pim-1 | | Authors: | Mechin, I, Zhang, Y, Wang, R, Batchelor, J.D, Mclean, L. | | Deposit date: | 2016-11-07 | | Release date: | 2017-10-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.948 Å) | | Cite: | Discovery of N-substituted 7-azaindoles as PIM1 kinase inhibitors - Part I.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5TOE
 
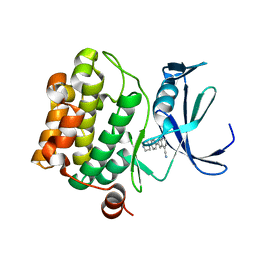 | | Pim-1 kinase in complex with a 7-azaindole | | Descriptor: | 2-[4-(piperazin-1-yl)phenyl]-1H-pyrrolo[2,3-b]pyridine-4-carbonitrile, Serine/threonine-protein kinase pim-1 | | Authors: | Mclean, L, Mechin, I, Zhang, Y, Wang, R, Batchelor, J.D. | | Deposit date: | 2016-10-17 | | Release date: | 2017-10-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Discovery of N-substituted 7-azaindoles as PIM1 kinase inhibitors - Part I.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
7FIA
 
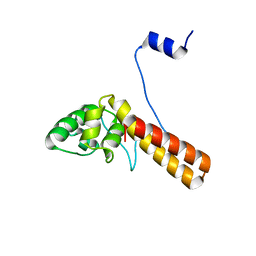 | | Structure of AcrIF23 | | Descriptor: | AcrIF23 | | Authors: | Ren, J, Yue, F. | | Deposit date: | 2021-07-30 | | Release date: | 2022-07-27 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural and mechanistic insights into the inhibition of type I-F CRISPR-Cas system by anti-CRISPR protein AcrIF23.
J.Biol.Chem., 298, 2022
|
|
7BI9
 
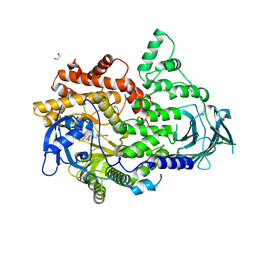 | | PI3KC2a core in complex with PIK90 | | Descriptor: | 1,2-ETHANEDIOL, N-(2,3-DIHYDRO-7,8-DIMETHOXYIMIDAZO[1,2-C] QUINAZOLIN-5-YL)NICOTINAMIDE, Phosphatidylinositol 4-phosphate 3-kinase C2 domain-containing subunit alpha,Phosphatidylinositol 4-phosphate 3-kinase C2 domain-containing subunit alpha | | Authors: | Lo, W.T, Roske, Y, Daumke, O, Haucke, V. | | Deposit date: | 2021-01-12 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis of phosphatidylinositol 3-kinase C2 alpha function.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7BI6
 
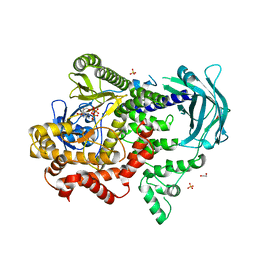 | | PI3KC2a core in complex with ATP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Lo, W.T, Roske, Y, Daumke, O, Haucke, V. | | Deposit date: | 2021-01-12 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis of phosphatidylinositol 3-kinase C2 alpha function.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7BI4
 
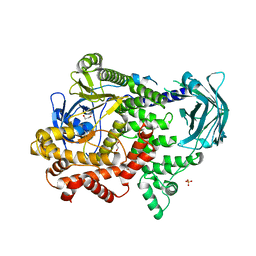 | | PI3KC2a core apo | | Descriptor: | 1,2-ETHANEDIOL, Phosphatidylinositol 4-phosphate 3-kinase C2 domain-containing subunit alpha,Phosphatidylinositol 4-phosphate 3-kinase C2 domain-containing subunit alpha, SULFATE ION | | Authors: | Lo, W.T, Roske, Y, Daumke, O, Haucke, V. | | Deposit date: | 2021-01-12 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural basis of phosphatidylinositol 3-kinase C2 alpha function.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7BI2
 
 | | PI3KC2aDeltaN and DeltaC-C2 | | Descriptor: | 1,2-ETHANEDIOL, 9-(6-aminopyridin-3-yl)-1-[3-(trifluoromethyl)phenyl]benzo[h][1,6]naphthyridin-2(1H)-one, IODIDE ION, ... | | Authors: | Lo, W.T, Roske, Y, Daumke, O, Haucke, V. | | Deposit date: | 2021-01-12 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structural basis of phosphatidylinositol 3-kinase C2 alpha function.
Nat.Struct.Mol.Biol., 29, 2022
|
|
8HQT
 
 | |
8HQW
 
 | |
8HQV
 
 | |
7W3Y
 
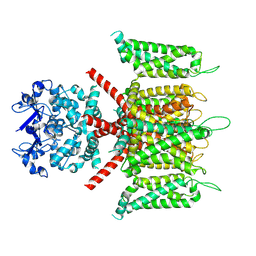 | | CryoEM structure of human Kv4.3 | | Descriptor: | Isoform 2 of Potassium voltage-gated channel subfamily D member 3 | | Authors: | Ma, D.M, Guo, J.T. | | Deposit date: | 2021-11-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for the gating modulation of Kv4.3 by auxiliary subunits.
Cell Res., 32, 2022
|
|
7W6S
 
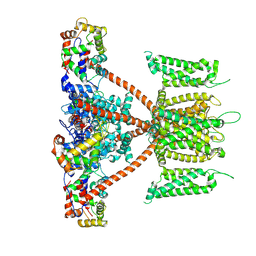 | | CryoEM structure of human KChIP2-Kv4.3 complex | | Descriptor: | Isoform 2 of Potassium voltage-gated channel subfamily D member 3, Kv channel-interacting protein 2 | | Authors: | Ma, D.M, Guo, J.T. | | Deposit date: | 2021-12-02 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for the gating modulation of Kv4.3 by auxiliary subunits.
Cell Res., 32, 2022
|
|
8HR0
 
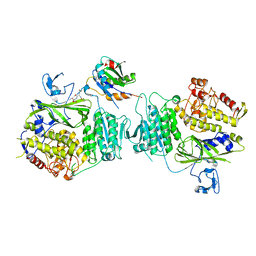 | | The complex structure of COPII coat with HCoV-OC43 DD sorting motif | | Descriptor: | HCoV-OC43, Protein transport protein Sec23A, Protein transport protein Sec24A, ... | | Authors: | Ma, W.F, Nan, Y.N, Yang, M.R, Li, Y.Q. | | Deposit date: | 2022-12-14 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | Tighter ER retention of SARS-CoV-2 Omicron spike caused by a constellation of folding disruptive mutations
To Be Published
|
|
7W6T
 
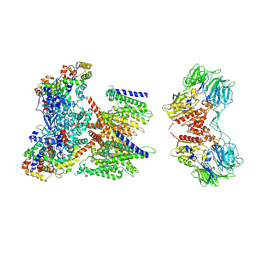 | | CryoEM structure of human KChIP1-Kv4.3-DPP6 complex | | Descriptor: | Dipeptidyl aminopeptidase-like protein 6, Isoform 2 of Potassium voltage-gated channel subfamily D member 3, Kv channel-interacting protein 1 | | Authors: | Ma, D.M, Guo, J.T. | | Deposit date: | 2021-12-02 | | Release date: | 2022-11-02 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Structural basis for the gating modulation of Kv4.3 by auxiliary subunits.
Cell Res., 32, 2022
|
|
7W6N
 
 | | CryoEM structure of human KChIP1-Kv4.3 complex | | Descriptor: | Isoform 2 of Potassium voltage-gated channel subfamily D member 3, Kv channel-interacting protein 1 | | Authors: | Ma, D.M, Guo, J.T. | | Deposit date: | 2021-12-02 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for the gating modulation of Kv4.3 by auxiliary subunits.
Cell Res., 32, 2022
|
|
8VH5
 
 | | Cryo-EM structure of Rab12-LRRK2 complex in the LRRK2 dimer state | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Leucine-rich repeat serine/threonine-protein kinase 2, MAGNESIUM ION, ... | | Authors: | Zhu, H, Sun, J. | | Deposit date: | 2023-12-30 | | Release date: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | RAB12-LRRK2 complex suppresses primary ciliogenesis and regulates centrosome homeostasis in astrocytes.
Nat Commun, 15, 2024
|
|
8VH4
 
 | | Cryo-EM structure of Rab12-LRRK2 complex in the LRRK2 monomer state | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Leucine-rich repeat serine/threonine-protein kinase 2, MAGNESIUM ION, ... | | Authors: | Zhu, H, Sun, J. | | Deposit date: | 2023-12-30 | | Release date: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | RAB12-LRRK2 complex suppresses primary ciliogenesis and regulates centrosome homeostasis in astrocytes.
Nat Commun, 15, 2024
|
|
8W71
 
 | | Structural basis of chorismate isomerization by Arabidopsis isochorismate synthase ICS1 | | Descriptor: | (3R,4R)-3-[(1-carboxyethenyl)oxy]-4-hydroxycyclohexa-1,5-diene-1-carboxylic acid, FORMIC ACID, Isochorismate synthase 1, ... | | Authors: | Su, Z.H, Ming, Z.H. | | Deposit date: | 2023-08-30 | | Release date: | 2024-05-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.121 Å) | | Cite: | Structural basis of chorismate isomerization by Arabidopsis ISOCHORISMATE SYNTHASE1.
Plant Physiol., 196, 2024
|
|
8W6V
 
 | |
5GIY
 
 | | HSA-Palmitic acid-[RuCl5(ind)]2- | | Descriptor: | PALMITIC ACID, Serum albumin, pentakis(chloranyl)-(1~{H}-indazol-2-ium-2-yl)ruthenium(1-) | | Authors: | Yang, F, Wang, T. | | Deposit date: | 2016-06-25 | | Release date: | 2017-07-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.543 Å) | | Cite: | Structure of HSA-Palmitic acid-[RuCl5(ind)]2-
To Be Published
|
|
8K36
 
 | |
8K35
 
 | | Structure of the bacteriophage lambda tail tip complex | | Descriptor: | IRON/SULFUR CLUSTER, Tail tip assembly protein I, Tail tip protein L, ... | | Authors: | Xiao, H, Tan, L, Cheng, L.P, Liu, H.R. | | Deposit date: | 2023-07-14 | | Release date: | 2023-11-15 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Structure of the siphophage neck-Tail complex suggests that conserved tail tip proteins facilitate receptor binding and tail assembly.
Plos Biol., 21, 2023
|
|
8K37
 
 | | Structure of the bacteriophage lambda neck | | Descriptor: | Head-tail connector protein FII, Tail tube protein, Tail tube terminator protein | | Authors: | Xiao, H, Tan, L, Cheng, L.P, Liu, H.R. | | Deposit date: | 2023-07-14 | | Release date: | 2023-11-15 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the siphophage neck-Tail complex suggests that conserved tail tip proteins facilitate receptor binding and tail assembly.
Plos Biol., 21, 2023
|
|
