4DSR
 
 | | Crystal structure of peroxiredoxin Ahp1 from Saccharomyces cerevisiae in reduced form | | Descriptor: | Peroxiredoxin type-2 | | Authors: | Lian, F.M, Yu, J, Ma, X.X, Yu, X.J, Chen, Y, Zhou, C.Z. | | Deposit date: | 2012-02-19 | | Release date: | 2012-04-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Structural Snapshots of Yeast Alkyl Hydroperoxide Reductase Ahp1 Peroxiredoxin Reveal a Novel Two-cysteine Mechanism of Electron Transfer to Eliminate Reactive Oxygen Species.
J.Biol.Chem., 287, 2012
|
|
4DE2
 
 | | CTX-M-9 class A beta-lactamase complexed with compound 12 | | Descriptor: | 3-[(dimethylamino)methyl]-N-[3-(1H-tetrazol-5-yl)phenyl]benzamide, Beta-lactamase, DIMETHYL SULFOXIDE | | Authors: | Nichols, D.A, Chen, Y. | | Deposit date: | 2012-01-19 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-Based Design of Potent and Ligand-Efficient Inhibitors of CTX-M Class A Beta-Lactamase
J.Med.Chem., 55, 2012
|
|
4DSQ
 
 | | Crystal structure of peroxiredoxin Ahp1 from Saccharomyces cerevisiae in oxidized form | | Descriptor: | Peroxiredoxin type-2 | | Authors: | Lian, F.M, Yu, J, Ma, X.X, Yu, X.J, Chen, Y, Zhou, C.Z. | | Deposit date: | 2012-02-19 | | Release date: | 2012-04-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Snapshots of Yeast Alkyl Hydroperoxide Reductase Ahp1 Peroxiredoxin Reveal a Novel Two-cysteine Mechanism of Electron Transfer to Eliminate Reactive Oxygen Species
J.Biol.Chem., 287, 2012
|
|
4DE1
 
 | | CTX-M-9 class A beta-lactamase complexed with compound 18 | | Descriptor: | Beta-lactamase, DIMETHYL SULFOXIDE, N-[3-(1H-tetrazol-5-yl)phenyl]-2H-indazole-5-carboxamide | | Authors: | Nichols, D.A, Chen, Y. | | Deposit date: | 2012-01-19 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Structure-Based Design of Potent and Ligand-Efficient Inhibitors of CTX-M Class A Beta-Lactamase
J.Med.Chem., 55, 2012
|
|
2KGP
 
 | | Structural basis for stabilization of the tau pre-mRNA splicing regulatory element by Novantrone (Mitoxantrone) | | Descriptor: | 1,4-DIHYDROXY-5,8-BIS({2-[(2-HYDROXYETHYL)AMINO]ETHYL}AMINO)-9,10-ANTHRACENEDIONE, RNA (25-MER) | | Authors: | Zheng, S, Chen, Y, Donahue, C.P, Wolfe, M.S, Varani, G. | | Deposit date: | 2009-03-13 | | Release date: | 2009-06-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for stabilization of the tau pre-mRNA splicing regulatory element by novantrone (mitoxantrone).
Chem.Biol., 16, 2009
|
|
6VNU
 
 | | X-ray Crystal Structure of Ruthenocenyl-7-Aminocephalosporanic Acid Covalent Acyl-Enzyme Complex with CTX-M-14 E166A Beta-Lactamase | | Descriptor: | Beta-lactamase, POTASSIUM ION, [(1,2,3,4,5-eta)-1-(4-{[carboxy(4-carboxy-5-methylidene-5,6-dihydro-2H-1,3-thiazin-2-yl)methyl]amino}-4-oxobutanoyl)cyclopentadienyl][(1,2,3,4,5-eta)-cyclopentadienyl]ruthenium, ... | | Authors: | Lewandowski, E.M, Jacobs, L.M.C, Chen, Y. | | Deposit date: | 2020-01-29 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Metallocenyl 7-ACA Conjugates: Antibacterial Activity Studies and Atomic-Resolution X-ray Crystal Structure with CTX-M beta-Lactamase.
Chembiochem, 21, 2020
|
|
7STA
 
 | |
4V8X
 
 | | Structure of Thermus thermophilus ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Feng, S, Chen, Y, Kamada, K, Wang, H, Tang, K, Wang, M, Gao, Y.G. | | Deposit date: | 2013-07-19 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Yoeb-Ribosome Structure: A Canonical Rnase that Requires the Ribosome for its Specific Activity.
Nucleic Acids Res., 41, 2013
|
|
5XU1
 
 | | Structure of a non-canonical ABC transporter from Streptococcus pneumoniae R6 | | Descriptor: | ABC transporter ATP-binding protein, ABC transporter permeae, MAGNESIUM ION | | Authors: | Yang, H.B, Jiang, Y.L, Hou, W.T, Chen, M.T, Chen, Y, Zhou, C.Z. | | Deposit date: | 2017-06-22 | | Release date: | 2018-01-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of a MacAB-like efflux pump from Streptococcus pneumoniae.
Nat Commun, 9, 2018
|
|
5XU0
 
 | | Structure of the membrane fusion protein Spr0693 from Streptococcus pneumoniae R6 | | Descriptor: | Membrane-fusion protein | | Authors: | Yang, H.B, Jiang, Y.L, Hou, W.T, Chen, M.T, Chen, Y, Zhou, C.Z. | | Deposit date: | 2017-06-22 | | Release date: | 2018-01-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of a MacAB-like efflux pump from Streptococcus pneumoniae.
Nat Commun, 9, 2018
|
|
5XP7
 
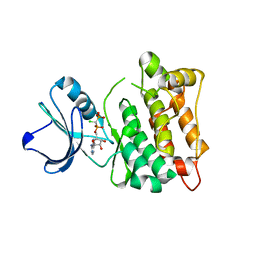 | | C-Src in complex with ATP-CHCl | | Descriptor: | GLYCEROL, MAGNESIUM ION, Proto-oncogene tyrosine-protein kinase Src, ... | | Authors: | Guo, M, Dai, S, Duan, Y, Chen, L, Chen, Y. | | Deposit date: | 2017-06-01 | | Release date: | 2017-09-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.012 Å) | | Cite: | Remarkably Stereospecific Utilization of ATP alpha , beta-Halomethylene Analogues by Protein Kinases.
J. Am. Chem. Soc., 139, 2017
|
|
5K5S
 
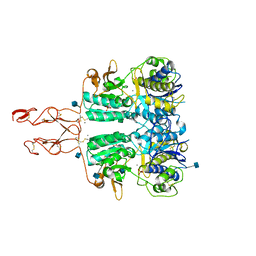 | | Crystal structure of the active form of human calcium-sensing receptor extracellular domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Extracellular calcium-sensing receptor, ... | | Authors: | Geng, Y, Mosyak, L, Kurinov, I, Zuo, H, Sturchler, E, Cheng, T.C, Subramanyam, P, Brown, A.P, Brennan, S.C, Mun, H.-C, Bush, M, Chen, Y, Nguyen, T, Cao, B, Chang, D, Quick, M, Conigrave, A, Colecraft, H.M, McDonald, P, Fan, Q.R. | | Deposit date: | 2016-05-23 | | Release date: | 2016-08-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural mechanism of ligand activation in human calcium-sensing receptor.
Elife, 5, 2016
|
|
5K5T
 
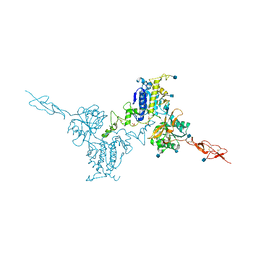 | | Crystal structure of the inactive form of human calcium-sensing receptor extracellular domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Extracellular calcium-sensing receptor, ... | | Authors: | Geng, Y, Mosyak, L, Kurinov, I, Zuo, H, Sturchler, E, Cheng, T.C, Subramanyam, P, Brown, A.P, Brennan, S.C, Mun, H.-C, Bush, M, Chen, Y, Nguyen, T, Cao, B, Chang, D, Quick, M, Conigrave, A, Colecraft, H.M, McDonald, P, Fan, Q.R. | | Deposit date: | 2016-05-23 | | Release date: | 2016-08-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural mechanism of ligand activation in human calcium-sensing receptor.
Elife, 5, 2016
|
|
7PKS
 
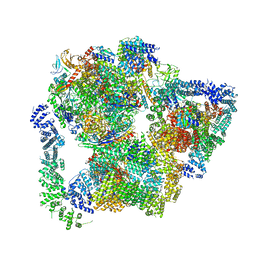 | | Structural basis of Integrator-mediated transcription regulation | | Descriptor: | DNA Template, DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, ... | | Authors: | Fianu, I, Chen, Y, Dienemann, C, Cramer, P. | | Deposit date: | 2021-08-26 | | Release date: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of Integrator-mediated transcription regulation.
Science, 374, 2021
|
|
4ZNQ
 
 | | Crystal structure of Dln1 complexed with Man(alpha1-2)Man | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Jia, N, Jiang, Y.L, Cheng, W, Wang, H.W, Zhou, C.Z, Chen, Y. | | Deposit date: | 2015-05-05 | | Release date: | 2016-01-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for receptor recognition and pore formation of a zebrafish aerolysin-like protein.
Embo Rep., 17, 2016
|
|
4ZNO
 
 | | Crystal structure of Dln1 complexed with sucrose | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, Natterin-like protein, ... | | Authors: | Jia, N, Jiang, Y.L, Cheng, W, Wang, H.W, Zhou, C.Z, Chen, Y. | | Deposit date: | 2015-05-05 | | Release date: | 2016-01-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural basis for receptor recognition and pore formation of a zebrafish aerolysin-like protein.
Embo Rep., 17, 2016
|
|
4ZNR
 
 | | Crystal structure of Dln1 complexed with Man(alpha1-3)Man | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Jia, N, Jiang, Y.L, Cheng, W, Wang, H.W, Zhou, C.Z, Chen, Y. | | Deposit date: | 2015-05-05 | | Release date: | 2016-01-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for receptor recognition and pore formation of a zebrafish aerolysin-like protein.
Embo Rep., 17, 2016
|
|
8HLL
 
 | | Crystal structure of p53/BCL2 fusion complex (complex 1) | | Descriptor: | Apoptosis regulator Bcl-2, Cellular tumor antigen p53, ZINC ION | | Authors: | Wei, H, Guo, M, Wang, H, Chen, Y. | | Deposit date: | 2022-11-30 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structures of p53/BCL-2 complex suggest a mechanism for p53 to antagonize BCL-2 activity.
Nat Commun, 14, 2023
|
|
8HLN
 
 | | Crystal structure of p53/BCL2 fusion complex(complex3) | | Descriptor: | Apoptosis regulator Bcl-2, Cellular tumor antigen p53, ZINC ION | | Authors: | Guo, M, Wei, H, Wang, H, Chen, Y. | | Deposit date: | 2022-11-30 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.354 Å) | | Cite: | Structures of p53/BCL-2 complex suggest a mechanism for p53 to antagonize BCL-2 activity.
Nat Commun, 14, 2023
|
|
8HLM
 
 | | Crystal structure of p53/BCL2 fusion complex (complex 2) | | Descriptor: | Apoptosis regulator Bcl-2, Cellular tumor antigen p53, ZINC ION | | Authors: | Guo, M, Wang, H, Wei, H, Chen, Y. | | Deposit date: | 2022-11-30 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.522 Å) | | Cite: | Structures of p53/BCL-2 complex suggest a mechanism for p53 to antagonize BCL-2 activity.
Nat Commun, 14, 2023
|
|
5LSW
 
 | | A CAF40-binding motif facilitates recruitment of the CCR4-NOT complex to mRNAs targeted by Drosophila Roquin | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cell differentiation protein RCD1 homolog, LD12033p, ... | | Authors: | Sgromo, A, Raisch, T, Bawankar, P, Bhandari, D, Chen, Y, Kuzuoglu-Ozturk, D, Weichenrieder, O, Izaurralde, E. | | Deposit date: | 2016-09-05 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A CAF40-binding motif facilitates recruitment of the CCR4-NOT complex to mRNAs targeted by Drosophila Roquin.
Nat Commun, 8, 2017
|
|
6Y9D
 
 | | Crystal structure of the quaternary ammonium Rieske monooxygenase CntA in complex with substrate L-Carnitine | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CARNITINE, Carnitine monooxygenase oxygenase subunit, ... | | Authors: | Quareshy, M, Shanmugam, M, Bugg, T.D, Cameron, A, Chen, Y. | | Deposit date: | 2020-03-06 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural basis of carnitine monooxygenase CntA substrate specificity, inhibition, and intersubunit electron transfer.
J.Biol.Chem., 296, 2020
|
|
5XVS
 
 | | Crystal structure of UDP-GlcNAc 2-epimerase NeuC complexed with UDP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GDP/UDP-N,N'-diacetylbacillosamine 2-epimerase (Hydrolyzing), LITHIUM ION, ... | | Authors: | Ko, T.P, Hsieh, T.J, Chen, S.C, Wu, S.C, Guan, H.H, Yang, C.H, Chen, C.J, Chen, Y. | | Deposit date: | 2017-06-28 | | Release date: | 2018-04-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.383 Å) | | Cite: | The tetrameric structure of sialic acid-synthesizing UDP-GlcNAc 2-epimerase fromAcinetobacter baumannii: A comparative study with human GNE.
J. Biol. Chem., 293, 2018
|
|
6Y8S
 
 | | Crystal structure of the quaternary ammonium Rieske monooxygenase CntA in complex with substrate gamma-butyrobetaine | | Descriptor: | 3-CARBOXY-N,N,N-TRIMETHYLPROPAN-1-AMINIUM, Carnitine monooxygenase oxygenase subunit, FE (III) ION, ... | | Authors: | Quareshy, M, Shanmugam, M, Bugg, T.D, Cameron, A, Chen, Y. | | Deposit date: | 2020-03-05 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.629 Å) | | Cite: | Structural basis of carnitine monooxygenase CntA substrate specificity, inhibition, and intersubunit electron transfer.
J.Biol.Chem., 296, 2020
|
|
6Y9C
 
 | | The structure of a quaternary ammonium Rieske monooxygenase reveals insights into carnitine oxidation by gut microbiota and inter-subunit electron transfer | | Descriptor: | CARNITINE, Carnitine monooxygenase oxygenase subunit, FE (III) ION, ... | | Authors: | Quareshy, M, Shanmugam, M, Bugg, T.D, Cameron, A, Chen, Y. | | Deposit date: | 2020-03-06 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterisation of an unusual cysteine pair in the Rieske carnitine monooxygenase CntA catalytic site.
Febs J., 2023
|
|
