8JU9
 
 | |
9AUK
 
 | | Structure of SARS-CoV-2 Mpro mutant (A173V) in complex with Nirmatrelvir (PF-07321332) | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Gajiwala, K.S, Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | Deposit date: | 2024-02-29 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | In vitro selection and analysis of SARS-CoV-2 nirmatrelvir resistance mutations contributing to clinical virus resistance surveillance.
Sci Adv, 10, 2024
|
|
9AUO
 
 | | Structure of SARS-CoV-2 Mpro mutant (L50F,T304I) | | Descriptor: | 3C-like proteinase nsp5 | | Authors: | Gajiwala, K.S, Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | Deposit date: | 2024-02-29 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.423 Å) | | Cite: | In vitro selection and analysis of SARS-CoV-2 nirmatrelvir resistance mutations contributing to clinical virus resistance surveillance.
Sci Adv, 10, 2024
|
|
9AUN
 
 | | Structure of SARS-CoV-2 Mpro mutant (T21I,T304I) | | Descriptor: | 3C-like proteinase nsp5 | | Authors: | Gajiwala, K.S, Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | Deposit date: | 2024-02-29 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | In vitro selection and analysis of SARS-CoV-2 nirmatrelvir resistance mutations contributing to clinical virus resistance surveillance.
Sci Adv, 10, 2024
|
|
9AUL
 
 | | Structure of SARS-CoV-2 Mpro mutant (A173V,T304I)) in complex with Nirmatrelvir (PF-07321332) | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Gajiwala, K.S, Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | Deposit date: | 2024-02-29 | | Release date: | 2024-08-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.421 Å) | | Cite: | In vitro selection and analysis of SARS-CoV-2 nirmatrelvir resistance mutations contributing to clinical virus resistance surveillance.
Sci Adv, 10, 2024
|
|
9AUJ
 
 | | Structure of SARS-CoV-2 Mpro mutant (S144A) in complex with Nirmatrelvir (PF-07321332) | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Gajiwala, K.S, Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | Deposit date: | 2024-02-29 | | Release date: | 2024-08-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.486 Å) | | Cite: | In vitro selection and analysis of SARS-CoV-2 nirmatrelvir resistance mutations contributing to clinical virus resistance surveillance.
Sci Adv, 10, 2024
|
|
9AUM
 
 | | Structure of SARS-CoV-2 Mpro mutant (T21I,S144A,T304I) in complex with Nirmatrelvir (PF-07321332) | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Gajiwala, K.S, Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | Deposit date: | 2024-02-29 | | Release date: | 2024-08-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.539 Å) | | Cite: | In vitro selection and analysis of SARS-CoV-2 nirmatrelvir resistance mutations contributing to clinical virus resistance surveillance.
Sci Adv, 10, 2024
|
|
5DDS
 
 | | Crystal structure of aminotransferase CrmG from Actinoalloteichus sp. WH1-2216-6 in complex with PLP | | Descriptor: | ACETIC ACID, CrmG, GLYCEROL, ... | | Authors: | Xu, J, Feng, Z, Liu, J. | | Deposit date: | 2015-08-25 | | Release date: | 2016-08-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Biochemical and Structural Insights into the Aminotransferase CrmG in Caerulomycin Biosynthesis
Acs Chem.Biol., 11, 2016
|
|
5DDW
 
 | | Crystal structure of aminotransferase CrmG from Actinoalloteichus sp. WH1-2216-6 in complex with the PMP external aldimine adduct with Caerulomycin M | | Descriptor: | CrmG, GLYCEROL, [5-hydroxy-4-({(E)-[(4-hydroxy-2,2'-bipyridin-6-yl)methylidene]amino}methyl)-6-methylpyridin-3-yl]methyl dihydrogen phosphate | | Authors: | Xu, J, Feng, Z, Liu, J. | | Deposit date: | 2015-08-25 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Biochemical and Structural Insights into the Aminotransferase CrmG in Caerulomycin Biosynthesis
Acs Chem.Biol., 11, 2016
|
|
5DDU
 
 | | Crystal structure of aminotransferase CrmG from Actinoalloteichus sp. WH1-2216-6 in complex with PMP | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, CrmG, GLYCEROL, ... | | Authors: | Xu, J, Feng, Z, Liu, J. | | Deposit date: | 2015-08-25 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Biochemical and Structural Insights into the Aminotransferase CrmG in Caerulomycin Biosynthesis
Acs Chem.Biol., 11, 2016
|
|
5I1V
 
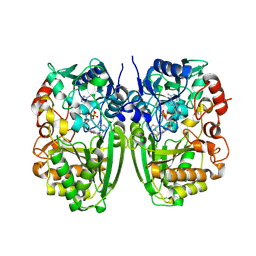 | |
5I1W
 
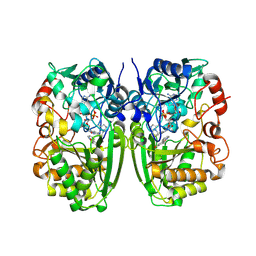 | | Crystal structure of CrmK, a flavoenzyme involved in the shunt product recycling mechanism in caerulomycin biosynthesis | | Descriptor: | 4-hydroxy[2,2'-bipyridine]-6-carbaldehyde, 6-(hydroxymethyl)[2,2'-bipyridin]-4-ol, CrmK, ... | | Authors: | Picard, M.-E, Barma, J, Shi, R. | | Deposit date: | 2016-02-07 | | Release date: | 2017-02-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Biochemical and structural insights into flavoenzyme CrmK reveals a shunt product recycling mechanism in caerulomycin biosynthesis
to be published
|
|
6J9M
 
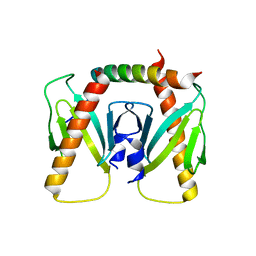 | | NmeBH+AcrIIC2 | | Descriptor: | AcrIIC2, CRISPR-associated endonuclease Cas9 | | Authors: | Zhu, Y.L, Gao, A, Serganov, A, Gao, P. | | Deposit date: | 2019-01-23 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.394 Å) | | Cite: | Diverse Mechanisms of CRISPR-Cas9 Inhibition by Type IIC Anti-CRISPR Proteins.
Mol. Cell, 74, 2019
|
|
6J9K
 
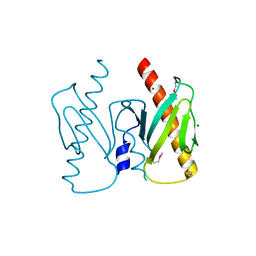 | | Apo-AcrIIC2 | | Descriptor: | AcrIIC2, MAGNESIUM ION | | Authors: | Zhu, Y.L, Gao, A, Serganov, A, Gao, P. | | Deposit date: | 2019-01-23 | | Release date: | 2019-03-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.234 Å) | | Cite: | Diverse Mechanisms of CRISPR-Cas9 Inhibition by Type IIC Anti-CRISPR Proteins.
Mol. Cell, 74, 2019
|
|
6J9L
 
 | | FnoBH+AcrIIC2 | | Descriptor: | AcrIIC2, HNH endonuclease family protein | | Authors: | Zhu, Y.L, Gao, A, Serganov, A, Gao, P. | | Deposit date: | 2019-01-23 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Diverse Mechanisms of CRISPR-Cas9 Inhibition by Type IIC Anti-CRISPR Proteins.
Mol. Cell, 74, 2019
|
|
6J9N
 
 | | NmeHNH+AcrIIC3 | | Descriptor: | AcrIIC3, CRISPR-associated endonuclease Cas9 | | Authors: | Zhu, Y.L, Gao, A, Serganov, A, Gao, P. | | Deposit date: | 2019-01-23 | | Release date: | 2019-03-06 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (2.606 Å) | | Cite: | Diverse Mechanisms of CRISPR-Cas9 Inhibition by Type IIC Anti-CRISPR Proteins.
Mol. Cell, 74, 2019
|
|
5UCA
 
 | |
5UC8
 
 | | Crystal structure of human Heme Oxygenase-2 | | Descriptor: | Heme oxygenase 2 | | Authors: | Luo, S, Tong, L. | | Deposit date: | 2016-12-22 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Heme Oxygenase 2 Binds Myristate to Regulate Retrovirus Assembly and TLR4 Signaling.
Cell Host Microbe, 21, 2017
|
|
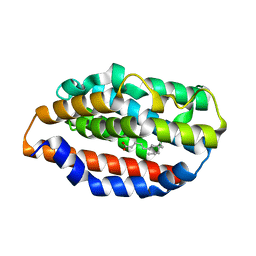
5UC9
 
 | |
8XOM
 
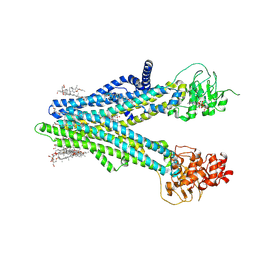 | | Cryo-EM structure of human ABCC4 in complex with ANP-bound in NBD1 and METHOTREXATE | | Descriptor: | 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, ATP-binding cassette sub-family C member 4, MAGNESIUM ION, ... | | Authors: | Zhang, P.F, Liu, Z. | | Deposit date: | 2024-01-01 | | Release date: | 2024-07-17 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | The ATP-bound inward-open conformation of ABCC4 reveals asymmetric ATP binding for substrate transport.
Febs Lett., 598, 2024
|
|
8XOL
 
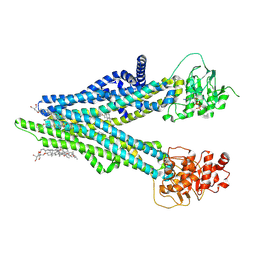 | | Cryo-EM structure of human ABCC4 with ANP bound in NBD1 | | Descriptor: | 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, ATP-binding cassette sub-family C member 4, MAGNESIUM ION, ... | | Authors: | Zhang, P.F, Liu, Z. | | Deposit date: | 2024-01-01 | | Release date: | 2024-07-17 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | The ATP-bound inward-open conformation of ABCC4 reveals asymmetric ATP binding for substrate transport.
Febs Lett., 598, 2024
|
|
8XOK
 
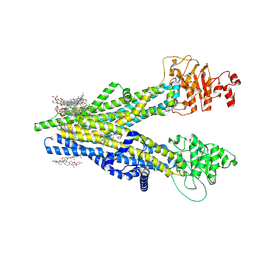 | | Cryo-EM structure of human ABCC4 | | Descriptor: | 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, ATP-binding cassette sub-family C member 4, PALMITIC ACID | | Authors: | Zhang, P.F, Liu, Z. | | Deposit date: | 2024-01-01 | | Release date: | 2024-07-24 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | The ATP-bound inward-open conformation of ABCC4 reveals asymmetric ATP binding for substrate transport.
Febs Lett., 598, 2024
|
|
5IHS
 
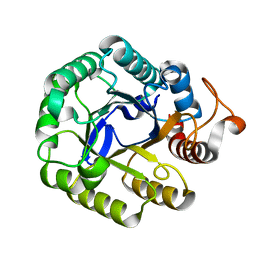 | | Structure of CHU_2103 from Cytophaga hutchinsonii | | Descriptor: | Endoglucanase, glycoside hydrolase family 5 protein | | Authors: | Silvaggi, N.R, Han, L. | | Deposit date: | 2016-02-29 | | Release date: | 2016-06-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Periplasmic Cytophaga hutchinsonii Endoglucanases Are Required for Use of Crystalline Cellulose as the Sole Source of Carbon and Energy.
Appl.Environ.Microbiol., 82, 2016
|
|
5YOW
 
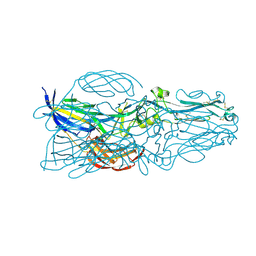 | | The post-fusion structure of the Heartland virus Gc glycoprotein | | Descriptor: | Glycoprotein polyprotein, alpha-L-fucopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)][alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Wu, Y, Gao, F, Qi, J.X, Chai, Y. | | Deposit date: | 2017-10-31 | | Release date: | 2017-11-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Postfusion Structure of the Heartland Virus Gc Glycoprotein Supports Taxonomic Separation of the Bunyaviral Families Phenuiviridae and Hantaviridae.
J. Virol., 92, 2018
|
|
5YC0
 
 | | Crystal structure of LP-46/N44 | | Descriptor: | Envelope glycoprotein, LP-46 | | Authors: | Zhang, X, Wang, X, He, Y. | | Deposit date: | 2017-09-05 | | Release date: | 2018-02-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Exceptional potency and structural basis of a T1249-derived lipopeptide fusion inhibitor against HIV-1, HIV-2, and simian immunodeficiency virus
J. Biol. Chem., 293, 2018
|
|
