8E5A
 
 | | Human L-type voltage-gated calcium channel Cav1.3 treated with 1.4 mM Sofosbuvir at 3.3 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gao, S, Yao, X, Yan, N. | | Deposit date: | 2022-08-20 | | Release date: | 2022-12-07 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for the severe adverse interaction of sofosbuvir and amiodarone on L-type Ca v channels.
Cell, 185, 2022
|
|
8E56
 
 | | Rabbit L-type voltage-gated calcium channel Cav1.1 in the presence of Amiodarone at 2.8 Angstrom resolution | | Descriptor: | (2-butyl-1-benzofuran-3-yl){4-[2-(diethylamino)ethoxy]-3,5-diiodophenyl}methanone, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gao, S, Yao, X, Yan, N. | | Deposit date: | 2022-08-20 | | Release date: | 2022-12-07 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for the severe adverse interaction of sofosbuvir and amiodarone on L-type Ca v channels.
Cell, 185, 2022
|
|
8E58
 
 | | Rabbit L-type voltage-gated calcium channel Cav1.1 in the presence of Amiodarone and 1 mM MNI-1 at 3.0 Angstrom resolution | | Descriptor: | (2-butyl-1-benzofuran-3-yl){4-[2-(diethylamino)ethoxy]-3,5-diiodophenyl}methanone, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gao, S, Yao, X, Yan, N. | | Deposit date: | 2022-08-20 | | Release date: | 2022-12-07 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for the severe adverse interaction of sofosbuvir and amiodarone on L-type Ca v channels.
Cell, 185, 2022
|
|
4NX2
 
 | | Crystal structure of DCYRS complexed with DCY | | Descriptor: | 3,5-dichloro-L-tyrosine, Tyrosine--tRNA ligase | | Authors: | Wang, J, Gong, W, Li, J, Gao, F, Li, H. | | Deposit date: | 2013-12-08 | | Release date: | 2014-09-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Significant expansion of fluorescent protein sensing ability through the genetic incorporation of superior photo-induced electron-transfer quenchers.
J.Am.Chem.Soc., 136, 2014
|
|
5GXO
 
 | | Discovery of a compound that activates SIRT3 to deacetylate Manganese Superoxide Dismutase | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase [Mn], mitochondrial | | Authors: | Lu, J, Li, J, Wu, M, Wang, J, Xia, Q. | | Deposit date: | 2016-09-19 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A small molecule activator of SIRT3 promotes deacetylation and activation of manganese superoxide dismutase.
Free Radic. Biol. Med., 112, 2017
|
|
4NXE
 
 | | Crystal structure of iLOV-I486(2LT) at pH 6.5 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin-2 | | Authors: | Wang, J, Liu, X, Li, J. | | Deposit date: | 2013-12-09 | | Release date: | 2014-09-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Significant expansion of fluorescent protein sensing ability through the genetic incorporation of superior photo-induced electron-transfer quenchers.
J.Am.Chem.Soc., 136, 2014
|
|
8EEA
 
 | | Structure of E.coli Septu (PtuAB) complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, PtuA, PtuB | | Authors: | Shen, Z.F, Fu, T.M. | | Deposit date: | 2022-09-06 | | Release date: | 2023-12-27 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | PtuA and PtuB assemble into an inflammasome-like oligomer for anti-phage defense.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8EE7
 
 | |
8EE4
 
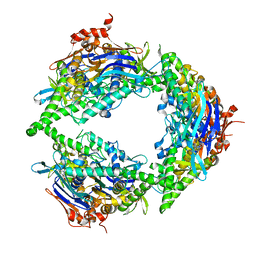 | | Structure of PtuA | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, PtuA | | Authors: | Shen, Z.F, Fu, T.M. | | Deposit date: | 2022-09-06 | | Release date: | 2024-01-03 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | PtuA and PtuB assemble into an inflammasome-like oligomer for anti-phage defense.
Nat.Struct.Mol.Biol., 31, 2024
|
|
4N9F
 
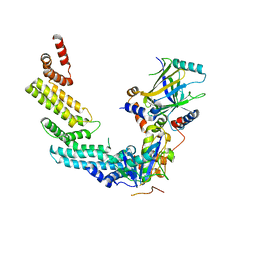 | | Crystal structure of the Vif-CBFbeta-CUL5-ElOB-ElOC pentameric complex | | Descriptor: | Core-binding factor subunit beta, Cullin-5, Transcription elongation factor B polypeptide 1, ... | | Authors: | Guo, Y.Y, Dong, L.Y, Huang, Z.W. | | Deposit date: | 2013-10-21 | | Release date: | 2014-01-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for hijacking CBF-b and CUL5 E3 ligase complex by HIV-1 Vif
Nature, 505, 2014
|
|
4NXG
 
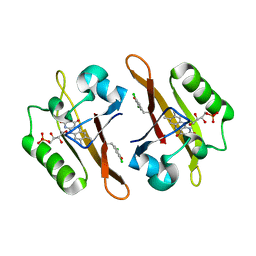 | | Crystal structure of iLOV-I486z(2LT) at pH 9.0 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin-2 | | Authors: | Wang, J, Liu, X, Li, J. | | Deposit date: | 2013-12-09 | | Release date: | 2014-09-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Significant expansion of fluorescent protein sensing ability through the genetic incorporation of superior photo-induced electron-transfer quenchers.
J.Am.Chem.Soc., 136, 2014
|
|
8EPL
 
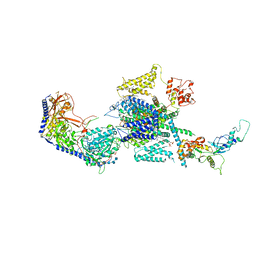 | | Human R-type voltage-gated calcium channel Cav2.3 at 3.1 Angstrom resolution | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gao, S, Yao, X, Yan, N. | | Deposit date: | 2022-10-06 | | Release date: | 2022-12-14 | | Last modified: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of the R-type human Ca v 2.3 channel reveal conformational crosstalk of the intracellular segments.
Nat Commun, 13, 2022
|
|
8EPM
 
 | | Human R-type voltage-gated calcium channel Cav2.3 CH2II-deleted mutant at 3.1 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gao, S, Yao, X, Yan, N. | | Deposit date: | 2022-10-06 | | Release date: | 2022-12-14 | | Last modified: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of the R-type human Ca v 2.3 channel reveal conformational crosstalk of the intracellular segments.
Nat Commun, 13, 2022
|
|
4NXB
 
 | | Crystal structure of iLOV-I486(2LT) at pH 7.0 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin-2 | | Authors: | Wang, J, Li, J, Liu, X. | | Deposit date: | 2013-12-09 | | Release date: | 2014-09-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.561 Å) | | Cite: | Significant expansion of fluorescent protein sensing ability through the genetic incorporation of superior photo-induced electron-transfer quenchers.
J.Am.Chem.Soc., 136, 2014
|
|
3HDZ
 
 | | Identification, Synthesis, and SAR of Amino Substituted Pyrido[3,2b]pryaziones as Potent and Selective PDE5 Inhibitors | | Descriptor: | 5-amino-1-butyl-7-phenyl-1,6-naphthyridin-4(1H)-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Cubbage, J.W, Brown, D.G, Jacobsen, E.J, Walker, J.K, Hughes, R.O. | | Deposit date: | 2009-05-07 | | Release date: | 2009-07-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification, synthesis and SAR of amino substituted pyrido[3,2b]pyrazinones as potent and selective PDE5 inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
8J4A
 
 | | Crystal structure of OY phytoplasma SAP05 in complex with AtRPN10 | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 4 homolog, Sequence-variable mosaic (SVM) signal sequence domain-containing protein | | Authors: | Dong, C, Yan, X, Yuan, X. | | Deposit date: | 2023-04-19 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Molecular basis of SAP05-mediated ubiquitin-independent proteasomal degradation of transcription factors.
Nat Commun, 15, 2024
|
|
8J4B
 
 | | Crystal structure of OY phytoplasma SAP05 in complex with AtSPL13 | | Descriptor: | Sequence-variable mosaic (SVM) signal sequence domain-containing protein, Squamosa promoter-binding-like protein 13A, ZINC ION | | Authors: | Dong, C, Yan, X, Yuan, X. | | Deposit date: | 2023-04-19 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis of SAP05-mediated ubiquitin-independent proteasomal degradation of transcription factors.
Nat Commun, 15, 2024
|
|
8J49
 
 | | Crystal structure of OY phytoplasma SAP05 in complex with AtSPL5 | | Descriptor: | Sequence-variable mosaic (SVM) signal sequence domain-containing protein, Squamosa promoter-binding-like protein 5, ZINC ION | | Authors: | Dong, C, Yan, X, Yuan, X. | | Deposit date: | 2023-04-19 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Molecular basis of SAP05-mediated ubiquitin-independent proteasomal degradation of transcription factors.
Nat Commun, 15, 2024
|
|
8J48
 
 | | Crystal structure of OY phytoplasma SAP05 in complex with AtGATA18 | | Descriptor: | GATA transcription factor 18, Sequence-variable mosaic (SVM) signal sequence domain-containing protein, ZINC ION | | Authors: | Dong, C, Yan, X, Yuan, X. | | Deposit date: | 2023-04-19 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Molecular basis of SAP05-mediated ubiquitin-independent proteasomal degradation of transcription factors.
Nat Commun, 15, 2024
|
|
8WRZ
 
 | | Cry-EM structure of cannabinoid receptor-arrestin 2 complex | | Descriptor: | (6~{a}~{R},9~{R},10~{a}~{R})-9-(hydroxymethyl)-3-(8-isothiocyanato-2-methyl-octan-2-yl)-6,6-dimethyl-6~{a},7,8,9,10,10~{a}-hexahydrobenzo[c]chromen-1-ol, Beta-arrestin-1, Soluble cytochrome b562,Cannabinoid receptor 1, ... | | Authors: | Wang, Y.X, Wang, T, Wu, L.J, Hua, T, Liu, Z.J. | | Deposit date: | 2023-10-16 | | Release date: | 2024-02-28 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of cannabinoid receptor CB1-beta-arrestin complex.
Protein Cell, 15, 2024
|
|
7SSB
 
 | | Co-structure of PKG1 regulatory domain with compound 33 | | Descriptor: | 4-({(2S,3S)-3-[(1S)-1-(3,5-dichlorophenyl)-2-hydroxyethoxy]-2-phenylpiperidin-1-yl}methyl)-3-nitrobenzoic acid, cGMP-dependent protein kinase 1 | | Authors: | Fischmann, T.O. | | Deposit date: | 2021-11-10 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Optimization and Mechanistic Investigations of Novel Allosteric Activators of PKG1 alpha.
J.Med.Chem., 65, 2022
|
|
7TRL
 
 | | Crystal structure of human BIRC2 BIR3 domain in complex with histone H3 | | Descriptor: | 1,2-ETHANEDIOL, Baculoviral IAP repeat-containing protein 2, Histone H3, ... | | Authors: | Klein, B.J, Tencer, A.H, Kutateladze, T.G. | | Deposit date: | 2022-01-29 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Molecular basis for nuclear accumulation and targeting of the inhibitor of apoptosis BIRC2.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7TRM
 
 | | Crystal structure of human BIRC2 BIR3 domain in complex with inhibitor LCL-161 | | Descriptor: | 1,2-ETHANEDIOL, Baculoviral IAP repeat-containing protein 2, LCL-161, ... | | Authors: | Tencer, A.H, Klein, B.J, Kutateladze, T.G. | | Deposit date: | 2022-01-29 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for nuclear accumulation and targeting of the inhibitor of apoptosis BIRC2.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7T31
 
 | | X-ray Structure of Clostridiodies difficile PilW | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Putative pilin protein chimera | | Authors: | Ronish, L.A, Piepenbrink, K.H. | | Deposit date: | 2021-12-06 | | Release date: | 2022-10-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Recognition of extracellular DNA by type IV pili promotes biofilm formation by Clostridioides difficile.
J.Biol.Chem., 298, 2022
|
|
3BTU
 
 | |
