6EO7
 
 | | X-ray structure of the complex between human alpha-thrombin and modified 15-mer DNA aptamer containing 5-(3-(acetamide-N-yl)-1-propen-1-yl)-2'-deoxyuridine residue | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, GA68B2 - MODIFIED HUMAN THROMBIN BINDING APTAMER, ... | | Authors: | Dolot, R.M, Nawrot, B, Yang, X. | | Deposit date: | 2017-10-09 | | Release date: | 2017-10-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Crystal structures of thrombin in complex with chemically modified thrombin DNA aptamers reveal the origins of enhanced affinity.
Nucleic Acids Res., 46, 2018
|
|
7CWK
 
 | |
5XGJ
 
 | | Crystal structure of PI3K complex with an inhibitor | | Descriptor: | 3-(4-morpholin-4-ylfuro[3,2-d]pyrimidin-2-yl)-5-[(phenylmethyl)amino]phenol, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, ... | | Authors: | Song, K, Yang, X, Zhao, Y, Jian, Z. | | Deposit date: | 2017-04-13 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Crystal structure of PI3K complex with an inhibitor
To Be Published
|
|
5XJO
 
 | | Plant receptor ERL1-TMM in complex with peptide EPF1 | | Descriptor: | LRR receptor-like serine/threonine-protein kinase ERL1, Protein EPIDERMAL PATTERNING FACTOR 1, Protein TOO MANY MOUTHS | | Authors: | Chai, J, Lin, G, Zhang, L, Han, Z, Shpak, E.D, Yang, X. | | Deposit date: | 2017-05-03 | | Release date: | 2019-01-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.626 Å) | | Cite: | A receptor-like protein acts as a specificity switch for the regulation of stomatal development.
Genes Dev., 31, 2017
|
|
7VWK
 
 | | The product template domain of AviM | | Descriptor: | Polyketide synthase | | Authors: | Feng, Y, Yang, X, Zheng, J. | | Deposit date: | 2021-11-10 | | Release date: | 2022-06-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Streptomyces viridochromogenes product template domain represents an evolutionary intermediate between dehydratase and aldol cyclase of type I polyketide synthases.
Commun Biol, 5, 2022
|
|
7V9G
 
 | | Native BEN4 domain of protein Bend3 with DNA | | Descriptor: | BEN domain-containing protein 3, DNA (5'-D(*GP*CP*AP*CP*CP*GP*CP*GP*TP*GP*GP*GP*GP*CP*CP*A)-3'), DNA (5'-D(*TP*GP*GP*CP*CP*CP*CP*AP*CP*GP*CP*GP*GP*TP*GP*C)-3') | | Authors: | Zhang, J, Zhang, Y, You, Q, Huang, C, Zhang, T, Wang, M, Zhang, T, Yang, X, Xiong, J, Li, Y, Liu, C.P, Zhang, Z, Xu, R.M, Zhu, B. | | Deposit date: | 2021-08-25 | | Release date: | 2022-02-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Highly enriched BEND3 prevents the premature activation of bivalent genes during differentiation.
Science, 375, 2022
|
|
7V9I
 
 | | The Monomer mutant of BEN4 domain of protein Bend3 with DNA | | Descriptor: | BEN domain-containing protein 3, DNA (5'-D(*AP*CP*CP*GP*CP*GP*TP*GP*GP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*CP*CP*AP*CP*GP*CP*GP*GP*T)-3') | | Authors: | Zhang, J, Zhang, Y, You, Q, Huang, C, Zhang, T, Wang, M, Zhang, T, Yang, X, Xiong, J, Li, Y, Liu, C.P, Zhang, Z, Xu, R.M, Zhu, B. | | Deposit date: | 2021-08-25 | | Release date: | 2022-02-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Highly enriched BEND3 prevents the premature activation of bivalent genes during differentiation.
Science, 375, 2022
|
|
7VGF
 
 | | cryo-EM structure of AMP-PNP bound human ABCB7 | | Descriptor: | Iron-sulfur clusters transporter ABCB7, mitochondrial, MAGNESIUM ION, ... | | Authors: | Yan, Q, Yang, X, Shen, Y. | | Deposit date: | 2021-09-16 | | Release date: | 2022-02-16 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of AMP-PNP-bound human mitochondrial ATP-binding cassette transporter ABCB7.
J.Struct.Biol., 214, 2022
|
|
7V9H
 
 | | The BEN3 domain of protein Bend3 | | Descriptor: | BEN domain-containing protein 3 | | Authors: | Zhang, J, Zhang, Y, You, Q, Huang, C, Zhang, T, Wang, M, Zhang, T, Yang, X, Xiong, J, Li, Y, Liu, C.P, Zhang, Z, Xu, R.M, Zhu, B. | | Deposit date: | 2021-08-25 | | Release date: | 2022-02-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.692 Å) | | Cite: | Highly enriched BEND3 prevents the premature activation of bivalent genes during differentiation.
Science, 375, 2022
|
|
7V9F
 
 | | Selenomethionine mutant (L740Sem) of BEN4 domain of protein Bend3 with DNA | | Descriptor: | BEN domain-containing protein 3, CITRIC ACID, DNA (5'-D(*GP*CP*AP*CP*CP*GP*CP*GP*TP*GP*GP*GP*GP*CP*CP*A)-3'), ... | | Authors: | Zhang, J, Zhang, Y, You, Q, Huang, C, Zhang, T, Wang, M, Zhang, T, Yang, X, Xiong, J, Li, Y, Liu, C.P, Zhang, Z, Xu, R.M, Zhu, B. | | Deposit date: | 2021-08-25 | | Release date: | 2022-02-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Highly enriched BEND3 prevents the premature activation of bivalent genes during differentiation.
Science, 375, 2022
|
|
7Y37
 
 | |
5Y6N
 
 | | Zika virus helicase in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Helicase domain from Genome polyprotein, MANGANESE (II) ION | | Authors: | Yang, X.Y, Chen, C, Tian, H.L, Chi, H, Mu, Z.Y, Zhang, T.Q, Yang, K.L, Zhao, Q, Liu, X.H, Wang, Z.F, Ji, X.Y, Yang, H.T. | | Deposit date: | 2017-08-12 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.571 Å) | | Cite: | Mechanism of ATP hydrolysis by the Zika virus helicase.
FASEB J., 32, 2018
|
|
9JBR
 
 | | P5A-ATPase ATP13A1 D533N mutant | | Descriptor: | Endoplasmic reticulum transmembrane helix translocase | | Authors: | Li, Y, Liao, J. | | Deposit date: | 2024-08-27 | | Release date: | 2025-06-11 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | ATP13A1 engages SEC61 to facilitate substrate-specific translocation.
Sci Adv, 11, 2025
|
|
9JBX
 
 | | P5A-ATPase ATP13A1 in E1P state | | Descriptor: | Endoplasmic reticulum transmembrane helix translocase | | Authors: | Li, Y, Liao, J. | | Deposit date: | 2024-08-27 | | Release date: | 2025-06-11 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | ATP13A1 engages SEC61 to facilitate substrate-specific translocation.
Sci Adv, 11, 2025
|
|
9JBM
 
 | | P5A-ATPase ATP13A1 reconstructed in nanodiscs | | Descriptor: | Endoplasmic reticulum transmembrane helix translocase, Putative endogenous substrate | | Authors: | Li, Y, Liao, J. | | Deposit date: | 2024-08-27 | | Release date: | 2025-06-11 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.83 Å) | | Cite: | ATP13A1 engages SEC61 to facilitate substrate-specific translocation.
Sci Adv, 11, 2025
|
|
9JBZ
 
 | | P5A-ATPase ATP13A1 in E2P state | | Descriptor: | Endoplasmic reticulum transmembrane helix translocase | | Authors: | Li, Y, Liao, J. | | Deposit date: | 2024-08-27 | | Release date: | 2025-06-11 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | ATP13A1 engages SEC61 to facilitate substrate-specific translocation.
Sci Adv, 11, 2025
|
|
5D2L
 
 | |
7FEE
 
 | | Crystal structure of the allosteric modulator ZCZ011 binding to CP55940-bound cannabinoid receptor 1 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-[(1R,2R,5R)-5-hydroxy-2-(3-hydroxypropyl)cyclohexyl]-5-(2-methyloctan-2-yl)phenol, 6-methyl-3-[(1S)-2-nitro-1-thiophen-2-yl-ethyl]-2-phenyl-1H-indole, ... | | Authors: | Wang, X, Zhao, C, Shao, Z. | | Deposit date: | 2021-07-19 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular mechanism of allosteric modulation for the cannabinoid receptor CB1.
Nat.Chem.Biol., 18, 2022
|
|
7FCZ
 
 | | Crystal Structure of human RIPK1 kinase domain in complex with a novel inhibitor | | Descriptor: | N-[(3S)-7-(2-cyclopropylethynyl)-5-methyl-4-oxidanylidene-2,3-dihydro-1,5-benzoxazepin-3-yl]-5-(phenylmethyl)-4H-1,2,4-triazole-3-carboxamide, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Su, H.X, Xie, H, Nie, T.Q, Li, M.J, Xu, Y.C. | | Deposit date: | 2021-07-15 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Potent and Selective RIPK1 Inhibitors Targeting Dual-Pockets for the Treatment of Systemic Inflammatory Response Syndrome and Sepsis.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7FD0
 
 | | Crystal Structure of human RIPK1 kinase domain in complex with a novel inhibitor | | Descriptor: | N-[(3S)-5-methyl-7-[2-(oxan-4-yl)ethynyl]-4-oxidanylidene-2,3-dihydro-1,5-benzoxazepin-3-yl]-5-(phenylmethyl)-4H-1,2,4-triazole-3-carboxamide, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Su, H.X, Xie, H, Nie, T.Q, Li, M.J, Xu, Y.C. | | Deposit date: | 2021-07-15 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Potent and Selective RIPK1 Inhibitors Targeting Dual-Pockets for the Treatment of Systemic Inflammatory Response Syndrome and Sepsis.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
5Y6M
 
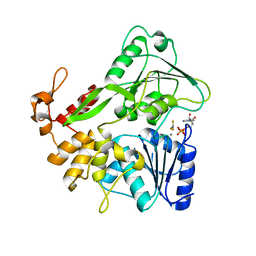 | | Zika virus helicase in complex with ADP-AlF3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Helicase domain from Genome polyprotein, ... | | Authors: | Yang, X.Y, Chen, C, Tian, H.L, Chi, H, Mu, Z.Y, Zhang, T.Q, Yang, K.L, Zhao, Q, Liu, X.H, Wang, Z.F, Ji, X.Y, Yang, H.T. | | Deposit date: | 2017-08-12 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Mechanism of ATP hydrolysis by the Zika virus helicase.
FASEB J., 32, 2018
|
|
4YNA
 
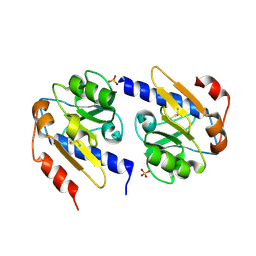 | | Oxidized YfiR | | Descriptor: | SULFATE ION, YfiR | | Authors: | Xu, M, Jiang, T. | | Deposit date: | 2015-03-09 | | Release date: | 2015-04-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structures of YfiR from Pseudomonas aeruginosa in two redox states
Biochem.Biophys.Res.Commun., 461, 2015
|
|
4YN9
 
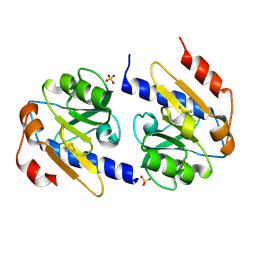 | | YfiR mutant-C110S | | Descriptor: | SULFATE ION, YfiR | | Authors: | Xu, M, Jiang, T. | | Deposit date: | 2015-03-09 | | Release date: | 2015-04-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structures of YfiR from Pseudomonas aeruginosa in two redox states
Biochem.Biophys.Res.Commun., 461, 2015
|
|
8X3L
 
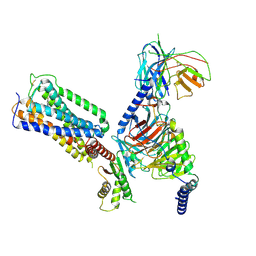 | | Cryo-EM structure of CB2-G protein complex | | Descriptor: | 4-[3-(4-fluoranyl-2~{H}-indazol-7-yl)-1,2,4-oxadiazol-5-yl]-1-(4-fluorophenyl)azepan-2-one, Cannabinoid receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Shen, S.Y, Yang, Z.Q, Shao, Z.H. | | Deposit date: | 2023-11-14 | | Release date: | 2024-11-20 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Entropy drives the ligand recognition in G-protein-coupled receptor subtypes.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
4Y21
 
 | | Crystal Structure of Munc13-1 MUN domain | | Descriptor: | Protein unc-13 homolog A | | Authors: | Yang, X.Y, Wang, S, Sheng, Y, Zhang, M, Zou, W.J, Wu, L.J, Kang, L.J, Rizo, J, Zhang, R.G, Xu, T, Ma, C. | | Deposit date: | 2015-02-09 | | Release date: | 2015-06-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Syntaxin opening by the MUN domain underlies the function of Munc13 in synaptic-vesicle priming.
Nat.Struct.Mol.Biol., 22, 2015
|
|
