7QD5
 
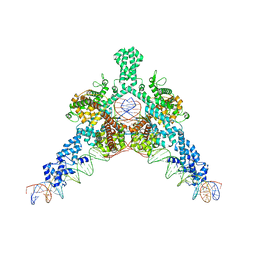 | | Cryo-EM structure of Tn4430 TnpA transposase from Tn3 family in complex with 48 bp long transposon end DNA | | Descriptor: | IR48 DNA substrate, non transferred strand, IR48 transferred strand, ... | | Authors: | Shkumatov, A.V, Oger, C.A, Aryanpour, N, Hallet, B.F, Efremov, R.G. | | Deposit date: | 2021-11-26 | | Release date: | 2022-10-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insight into Tn3 family transposition mechanism.
Nat Commun, 13, 2022
|
|
7PAG
 
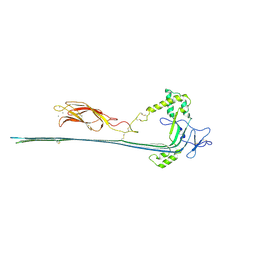 | | The pore conformation of lymphocyte perforin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Perforin-1 | | Authors: | Ivanova, M.E, Lukoyanova, N, Malhotra, S, Topf, M, Trapani, J.A, Voskoboinik, I, Saibil, H.R. | | Deposit date: | 2021-07-29 | | Release date: | 2022-02-16 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | The pore conformation of lymphocyte perforin.
Sci Adv, 8, 2022
|
|
2WKO
 
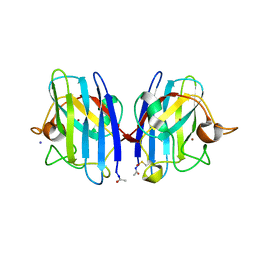 | | Structure of metal loaded Pathogenic SOD1 Mutant G93A. | | Descriptor: | COPPER (II) ION, IODIDE ION, SUPEROXIDE DISMUTASE [CU-ZN], ... | | Authors: | Antonyuk, S.V, Galaleldeen, A, Strange, R, Whitson, L, Narayana, N, Taylor, A, Schuermann, J.P, Holloway, S.P, Hasnain, S.S, Hart, P.J. | | Deposit date: | 2009-06-16 | | Release date: | 2009-11-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural and Biophysical Properties of Metal-Free Pathogenic Sod1 Mutants A4V and G93A.
Arch.Biochem.Biophys., 492, 2009
|
|
1L8J
 
 | | Crystal Structure of the Endothelial Protein C Receptor and Bound Phospholipid Molecule | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Endothelial protein C receptor, ... | | Authors: | Oganesyan, V, Oganesyan, N, Terzyan, S, Dongfeng, Q, Dauter, Z, Esmon, N.L, Esmon, C.T. | | Deposit date: | 2002-03-20 | | Release date: | 2002-06-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of the endothelial protein C receptor and a bound phospholipid.
J.Biol.Chem., 277, 2002
|
|
8U81
 
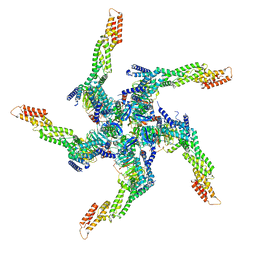 | | KCTD5/Cullin3/Gbeta1gamma2 Complex: State A From Composite RELION Multi-body Refinement Map | | Descriptor: | BTB/POZ domain-containing protein KCTD5, Cullin-3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Kuntz, D.A, Nguyen, D.M, Narayanan, N, Prive, G.G. | | Deposit date: | 2023-09-15 | | Release date: | 2023-10-11 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Structure and dynamics of a pentameric KCTD5/CUL3/G beta gamma E3 ubiquitin ligase complex.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8U7Z
 
 | | KCTD5/Cullin3/Gbeta1gamma2 Complex: Local Refinment of KCTD5(CTD)/Gbeta1gamma2 | | Descriptor: | BTB/POZ domain-containing protein KCTD5, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 | | Authors: | Kuntz, D.A, Nguyen, D.M, Narayanan, N, Prive, G.G. | | Deposit date: | 2023-09-15 | | Release date: | 2023-10-11 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structure and dynamics of a pentameric KCTD5/CUL3/G beta gamma E3 ubiquitin ligase complex.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8U82
 
 | | KCTD5/Cullin3/Gbeta1gamma2 Complex: State B From Composite RELION Multi-body Refinement Map | | Descriptor: | BTB/POZ domain-containing protein KCTD5, Cullin-3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Kuntz, D.A, Nguyen, D.M, Narayanan, N, Prive, G.G. | | Deposit date: | 2023-09-15 | | Release date: | 2023-10-11 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Structure and dynamics of a pentameric KCTD5/CUL3/G beta gamma E3 ubiquitin ligase complex.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8U80
 
 | | KCTD5/Cullin3/Gbeta1gamma2 Complex: Local Refinment of KCTD5(BTB)/Cullin3(NTD) | | Descriptor: | BTB/POZ domain-containing protein KCTD5, Cullin-3 | | Authors: | Kuntz, D.A, Nguyen, D.M, Narayanan, N, Prive, G.G. | | Deposit date: | 2023-09-15 | | Release date: | 2023-10-11 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure and dynamics of a pentameric KCTD5/CUL3/G beta gamma E3 ubiquitin ligase complex.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8U83
 
 | | KCTD5/Cullin3/Gbeta1gamma2 Complex: State C From Composite RELION Multi-body Refinement Map | | Descriptor: | BTB/POZ domain-containing protein KCTD5, Cullin-3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Kuntz, D.A, Nguyen, D.M, Narayanan, N, Prive, G.G. | | Deposit date: | 2023-09-15 | | Release date: | 2023-10-11 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.975 Å) | | Cite: | Structure and dynamics of a pentameric KCTD5/CUL3/G beta gamma E3 ubiquitin ligase complex.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8U84
 
 | | KCTD5/Cullin3/Gbeta1gamma2 Complex: State D From Composite RELION Multi-body Refinement Map | | Descriptor: | BTB/POZ domain-containing protein KCTD5, Cullin-3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Kuntz, D.A, Nguyen, D.M, Narayanan, N, Prive, G.G. | | Deposit date: | 2023-09-15 | | Release date: | 2023-10-11 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Structure and dynamics of a pentameric KCTD5/CUL3/G beta gamma E3 ubiquitin ligase complex.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
4R3U
 
 | | Crystal structure of 2-Hydroxyisobutyryl-CoA Mutase | | Descriptor: | 2-hydroxyisobutyryl-CoA mutase large subunit, 2-hydroxyisobutyryl-CoA mutase small subunit, 3-HYDROXYBUTANOYL-COENZYME A, ... | | Authors: | Zahn, M, Kurteva-Yaneva, N, Rohwerder, T, Straeter, N. | | Deposit date: | 2014-08-18 | | Release date: | 2015-03-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of the stereospecificity of bacterial B12-dependent 2-hydroxyisobutyryl-CoA mutase.
J.Biol.Chem., 290, 2015
|
|
5E5A
 
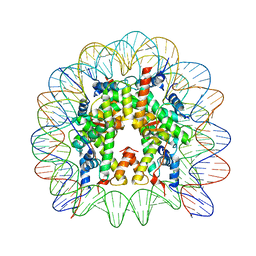 | | Crystal structure of the chromatin-tethering domain of Human cytomegalovirus IE1 protein bound to the nucleosome core particle | | Descriptor: | C-terminal domain of Regulatory protein IE1, DNA (146-MER), Histone H2A, ... | | Authors: | Fang, Q, Chen, P, Wang, M, Fang, J, Yang, N, Li, G, Xu, R.M. | | Deposit date: | 2015-10-08 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.809 Å) | | Cite: | Human cytomegalovirus IE1 protein alters the higher-order chromatin structure by targeting the acidic patch of the nucleosome
Elife, 5, 2016
|
|
5EX7
 
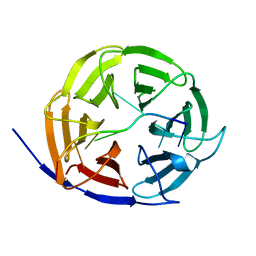 | | Crystal structure of Brat NHL domain in complex with an 8-nt hunchback mRNA | | Descriptor: | Brain tumor protein, RNA (5'-R(P*UP*UP*UP*GP*UP*UP*GP*U)-3') | | Authors: | Wang, Y, Yu, Z, Wang, M, Liu, C.P, Yang, N, Xu, R.M. | | Deposit date: | 2015-11-23 | | Release date: | 2015-12-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Brat NHL domain in complex with an 8-nt hunchback mRNA
To Be Published
|
|
5H1L
 
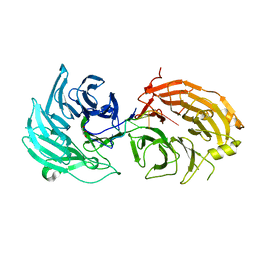 | | Crystal structure of WD40 repeat domains of Gemin5 in complex with 7-nt U4 snRNA fragment | | Descriptor: | GLYCEROL, Gem-associated protein 5, U4 snRNA (5'-R(*AP*UP*UP*UP*UP*UP*G)-3') | | Authors: | Jin, W, Wang, Y, Liu, C.P, Yang, N, Jin, M, Cong, Y, Wang, M, Xu, R.M. | | Deposit date: | 2016-10-10 | | Release date: | 2016-11-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for snRNA recognition by the double-WD40 repeat domain of Gemin5
Genes Dev., 30, 2016
|
|
5H1M
 
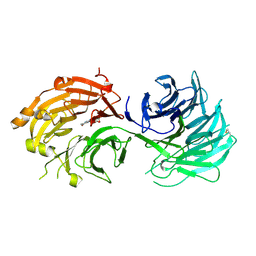 | | Crystal structure of WD40 repeat domains of Gemin5 in complex with M7G | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, Gem-associated protein 5 | | Authors: | Jin, W, Wang, Y, Liu, C.P, Yang, N, Jin, M, Cong, Y, Wang, M, Xu, R.M. | | Deposit date: | 2016-10-10 | | Release date: | 2016-11-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.492 Å) | | Cite: | Structural basis for snRNA recognition by the double-WD40 repeat domain of Gemin5
Genes Dev., 30, 2016
|
|
5C3I
 
 | | Crystal structure of the quaternary complex of histone H3-H4 heterodimer with chaperone ASF1 and the replicative helicase subunit MCM2 | | Descriptor: | DNA replication licensing factor MCM2,MCM2, Histone H3.1, Histone H4, ... | | Authors: | Wang, H, Wang, M, Yang, N, Xu, R.M. | | Deposit date: | 2015-06-17 | | Release date: | 2015-07-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of the quaternary complex of histone H3-H4 heterodimer with chaperone ASF1 and the replicative helicase subunit MCM2
Protein Cell, 6, 2015
|
|
5BTR
 
 | | Crystal structure of SIRT1 in complex with resveratrol and an AMC-containing peptide | | Descriptor: | AMC-containing peptide, NAD-dependent protein deacetylase sirtuin-1, RESVERATROL, ... | | Authors: | Cao, D, Wang, M, Qiu, X, Liu, D, Jiang, H, Yang, N, Xu, R.M. | | Deposit date: | 2015-06-03 | | Release date: | 2015-07-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for allosteric, substrate-dependent stimulation of SIRT1 activity by resveratrol
Genes Dev., 29, 2015
|
|
1T8B
 
 | | Crystal structure of refolded PHOU-like protein (gi 2983430) from Aquifex aeolicus | | Descriptor: | Phosphate transport system protein phoU homolog | | Authors: | Oganesyan, V, Kim, S.-H, Oganesyan, N, Jancarik, J, Adams, P.D, Kim, R, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-05-11 | | Release date: | 2004-12-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | Crystal structure of the "PhoU-like" phosphate uptake regulator from Aquifex aeolicus.
J.Bacteriol., 187, 2005
|
|
1T72
 
 | | Crystal structure of phosphate transport system protein phoU from Aquifex aeolicus | | Descriptor: | Phosphate transport system protein phoU homolog | | Authors: | Oganesyan, V, Kim, S.-H, Oganesyan, N, Jancarik, J, Adams, P.D, Kim, R, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-05-07 | | Release date: | 2004-12-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the "PhoU-like" phosphate uptake regulator from Aquifex aeolicus.
J.Bacteriol., 187, 2005
|
|
4PW7
 
 | | structure of UHRF2-SRA in complex with a 5mC-containing DNA | | Descriptor: | 5mC-containing DNA1, 5mC-containing DNA2, E3 ubiquitin-protein ligase UHRF2 | | Authors: | ZHou, T, Xiong, J, Wang, M, Yang, N, Wong, J, Zhu, B, Xu, R.M. | | Deposit date: | 2014-03-19 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural Basis for Hydroxymethylcytosine Recognition by the SRA Domain of UHRF2.
Mol.Cell, 54, 2014
|
|
4Q5W
 
 | | Crystal structure of extended-Tudor 9 of Drosophila melanogaster | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Maternal protein tudor | | Authors: | Ren, R, Liu, H, Wang, W, Wang, M, Yang, N, Dong, Y, Gong, W, Lehmann, R, Xu, R.M. | | Deposit date: | 2014-04-17 | | Release date: | 2014-05-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structure and domain organization of Drosophila Tudor
Cell Res., 24, 2014
|
|
4Q5Y
 
 | | Crystal structure of extended-Tudor 10-11 of Drosophila melanogaster | | Descriptor: | Maternal protein tudor | | Authors: | Liu, H, Ren, R, Wang, W, Wang, M, Yang, N, Dong, Y, Gong, W, Lehmann, R, Xu, R.M. | | Deposit date: | 2014-04-18 | | Release date: | 2014-05-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and domain organization of Drosophila Tudor
Cell Res., 24, 2014
|
|
4PW6
 
 | | structure of UHRF2-SRA in complex with a 5hmC-containing DNA, complex II | | Descriptor: | 5hmC-containing DNA1, 5hmC-containing DNA2, E3 ubiquitin-protein ligase UHRF2 | | Authors: | Zhou, T, Xiong, J, Wang, M, Yang, N, Wong, J, Zhu, B, Xu, R.M. | | Deposit date: | 2014-03-18 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.789 Å) | | Cite: | Structural Basis for Hydroxymethylcytosine Recognition by the SRA Domain of UHRF2.
Mol.Cell, 54, 2014
|
|
4PW5
 
 | | structure of UHRF2-SRA in complex with a 5hmC-containing DNA, complex I | | Descriptor: | 5hmC-containing DNA1, 5hmC-containing DNA2, E3 ubiquitin-protein ligase UHRF2 | | Authors: | ZHou, T, Xiong, J, Wang, M, Yang, N, Wong, J, Zhu, B, Xu, R.M. | | Deposit date: | 2014-03-18 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Structural Basis for Hydroxymethylcytosine Recognition by the SRA Domain of UHRF2.
Mol.Cell, 54, 2014
|
|
4KUL
 
 | | Crystal structure of N-terminal acetylated yeast Sir3 BAH domain V83P mutant | | Descriptor: | Regulatory protein SIR3 | | Authors: | Yang, D, Fang, Q, Wang, M, Ren, R, Wang, H, He, M, Sun, Y, Yang, N, Xu, R.M. | | Deposit date: | 2013-05-22 | | Release date: | 2013-08-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | N alpha-acetylated Sir3 stabilizes the conformation of a nucleosome-binding loop in the BAH domain.
Nat.Struct.Mol.Biol., 20, 2013
|
|
