8HGH
 
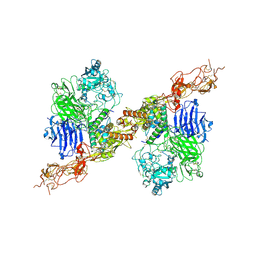 | | Structure of 2:2 PAPP-A.STC2 complex | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Pappalysin-1, Stanniocalcin-2, ZINC ION | | Authors: | Zhong, Q.H, Chu, H.L, Wang, G.P, Zhang, C, Wei, Y, Qiao, J, Hang, J. | | Deposit date: | 2022-11-14 | | Release date: | 2023-01-11 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.16 Å) | | Cite: | Structural insights into the covalent regulation of PAPP-A activity by proMBP and STC2.
Cell Discov, 8, 2022
|
|
8HGG
 
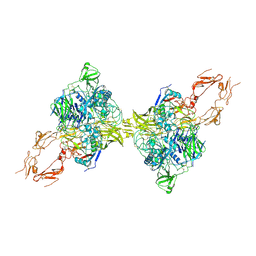 | | Structure of 2:2 PAPP-A.ProMBP complex | | Descriptor: | Bone marrow proteoglycan, Pappalysin-1, ZINC ION | | Authors: | Zhong, Q.H, Chu, H.L, Wang, G.P, Zhang, C, Wei, Y, Qiao, J, Hang, J. | | Deposit date: | 2022-11-14 | | Release date: | 2023-01-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Structural insights into the covalent regulation of PAPP-A activity by proMBP and STC2.
Cell Discov, 8, 2022
|
|
4ZIJ
 
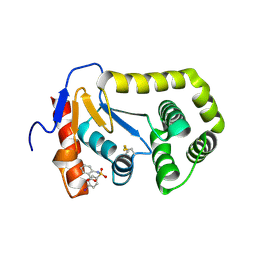 | | Crystal structure of E.Coli DsbA in complex with 2-(4-iodophenylsulfonamido) benzoic acid | | Descriptor: | 1,2-ETHANEDIOL, 2-{[(4-iodophenyl)sulfonyl]amino}benzoic acid, Thiol:disulfide interchange protein DsbA | | Authors: | Vazirani, M, Ilyichova, O.V, Scanlon, M.J. | | Deposit date: | 2015-04-28 | | Release date: | 2016-05-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Determination of ligand binding modes in weak protein-ligand complexes using sparse NMR data.
J.Biomol.Nmr, 66, 2016
|
|
6KLJ
 
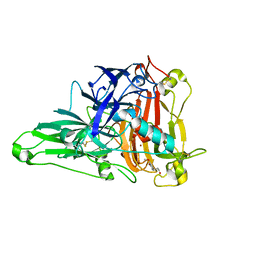 | | Crystal Structure of the Zea Mays laccase 3 complexed with coniferyl | | Descriptor: | (2E)-3-(4-hydroxy-3-methoxyphenyl)prop-2-enal, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Xie, T, Liu, Z.C, Wang, G.G. | | Deposit date: | 2019-07-30 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Structural basis for monolignol oxidation by a maize laccase.
Nat.Plants, 6, 2020
|
|
5ZLM
 
 | | Mutation in the trinuclear site of CotA-laccase: H491C mutant, PH 8.0 | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, GLYCEROL, ... | | Authors: | Xie, T, Liu, Z.C, Wang, G.G. | | Deposit date: | 2018-03-28 | | Release date: | 2018-05-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Insight into the Allosteric Coupling of Cu1 Site and Trinuclear Cu Cluster in CotA Laccase.
Chembiochem, 19, 2018
|
|
5ZLK
 
 | | Mutation in the trinuclear site of CotA-laccase: H493A mutant, PH 8.0 | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, GLYCEROL, ... | | Authors: | Xie, T, Liu, Z.C, Wang, G.G. | | Deposit date: | 2018-03-28 | | Release date: | 2018-05-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Insight into the Allosteric Coupling of Cu1 Site and Trinuclear Cu Cluster in CotA Laccase.
Chembiochem, 19, 2018
|
|
5ZLJ
 
 | | Crystal structure of CotA native enzyme, PH8.0 | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, GLYCEROL, ... | | Authors: | Xie, T, Liu, Z.C, Wang, G.G. | | Deposit date: | 2018-03-28 | | Release date: | 2018-05-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural Insight into the Allosteric Coupling of Cu1 Site and Trinuclear Cu Cluster in CotA Laccase.
Chembiochem, 19, 2018
|
|
8X3R
 
 | | Crystal structure of human WDR5 in complex with WDR5 | | Descriptor: | WD repeat-containing protein 5 | | Authors: | Liu, Y, Huang, X. | | Deposit date: | 2023-11-14 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | The NTE domain of PTEN alpha / beta promotes cancer progression by interacting with WDR5 via its SSSRRSS motif.
Cell Death Dis, 15, 2024
|
|
8X3S
 
 | | Crystal structure of human WDR5 in complex with PTEN | | Descriptor: | Phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN, WD repeat-containing protein 5 | | Authors: | Liu, Y, Huang, X, Shang, X. | | Deposit date: | 2023-11-14 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | The NTE domain of PTEN alpha / beta promotes cancer progression by interacting with WDR5 via its SSSRRSS motif.
Cell Death Dis, 15, 2024
|
|
6KLG
 
 | | Crystal Structure of the Zea Mays laccase 3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, ... | | Authors: | Xie, T, Liu, Z.C, Wang, G.G. | | Deposit date: | 2019-07-30 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for monolignol oxidation by a maize laccase.
Nat.Plants, 6, 2020
|
|
5ZLL
 
 | | Mutation in the trinuclear site of CotA-laccase: H493C mutant, PH 8.0 | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, GLYCEROL, ... | | Authors: | Xie, T, Liu, Z.C, Wang, G.G. | | Deposit date: | 2018-03-28 | | Release date: | 2018-05-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Insight into the Allosteric Coupling of Cu1 Site and Trinuclear Cu Cluster in CotA Laccase.
Chembiochem, 19, 2018
|
|
5X8H
 
 | | Crystal structure of the ketone reductase ChKRED20 from the genome of Chryseobacterium sp. CA49 | | Descriptor: | Short-chain dehydrogenase reductase | | Authors: | Zhao, F.J, Jin, Y, Liu, Z.C, Wang, G.G, Wu, Z.L. | | Deposit date: | 2017-03-02 | | Release date: | 2017-11-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure and iterative saturation mutagenesis of ChKRED20 for expanded catalytic scope
Appl. Microbiol. Biotechnol., 101, 2017
|
|
6KLI
 
 | | Crystal Structure of the Zea Mays laccase 3 complexed with sinapyl | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[(1E)-3-hydroxyprop-1-en-1-yl]-2,6-dimethoxyphenol, ... | | Authors: | Xie, T, Liu, Z.C, Wang, G.G. | | Deposit date: | 2019-07-30 | | Release date: | 2020-01-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for monolignol oxidation by a maize laccase.
Nat.Plants, 6, 2020
|
|
6LJJ
 
 | | Swine dUTPase in complex with alpha,beta-iminodUTP and magnesium ion | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, Deoxyuridine 5'-triphosphate nucleotidohydrolase, mitochondrial isoform 1, ... | | Authors: | Liang, R, Peng, G.Q. | | Deposit date: | 2019-12-16 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural comparisons of host and African swine fever virus dUTPases reveal new clues for inhibitor development.
J.Biol.Chem., 296, 2020
|
|
6BEA
 
 | |
6DNV
 
 | |
5DOO
 
 | | The structure of PKMT2 from Rickettsia typhi | | Descriptor: | CALCIUM ION, protein lysine methyltransferase 2 | | Authors: | Noinaj, N, Abeykoon, A, He, Y, Yang, D.C, Buchanan, S.K. | | Deposit date: | 2015-09-11 | | Release date: | 2016-08-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.133 Å) | | Cite: | Structural Insights into Substrate Recognition and Catalysis in Outer Membrane Protein B (OmpB) by Protein-lysine Methyltransferases from Rickettsia.
J.Biol.Chem., 291, 2016
|
|
5DO0
 
 | | The structure of PKMT1 from Rickettsia prowazekii | | Descriptor: | protein lysine methyltransferase 1 | | Authors: | Noinaj, N, Abeykoon, A, He, Y, Yang, D.C, Buchanan, S.K. | | Deposit date: | 2015-09-10 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Insights into Substrate Recognition and Catalysis in Outer Membrane Protein B (OmpB) by Protein-lysine Methyltransferases from Rickettsia.
J.Biol.Chem., 291, 2016
|
|
6DPS
 
 | |
5DNK
 
 | | The structure of PKMT1 from Rickettsia prowazekii in complex with AdoHcy | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, protein lysine methyltransferase 1 | | Authors: | Noinaj, N, Abeykoon, A, He, Y, Yang, D.C, Buchanan, S.K. | | Deposit date: | 2015-09-10 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insights into Substrate Recognition and Catalysis in Outer Membrane Protein B (OmpB) by Protein-lysine Methyltransferases from Rickettsia.
J.Biol.Chem., 291, 2016
|
|
5DPL
 
 | | The structure of PKMT2 from Rickettsia typhi in complex with AdoHcy | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, protein lysine methyltransferase 2 | | Authors: | Noinaj, N, Abeykoon, A, He, Y, Yang, D.C, Buchanan, S.K. | | Deposit date: | 2015-09-12 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Insights into Substrate Recognition and Catalysis in Outer Membrane Protein B (OmpB) by Protein-lysine Methyltransferases from Rickettsia.
J.Biol.Chem., 291, 2016
|
|
5DPD
 
 | | The structure of PKMT1 from Rickettsia prowazekii in complex with AdoMet | | Descriptor: | S-ADENOSYLMETHIONINE, protein lysine methyltransferase 1 | | Authors: | Noinaj, N, Abeykoon, A, He, Y, Yang, D.C, Buchanan, S.K. | | Deposit date: | 2015-09-12 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Insights into Substrate Recognition and Catalysis in Outer Membrane Protein B (OmpB) by Protein-lysine Methyltransferases from Rickettsia.
J.Biol.Chem., 291, 2016
|
|
6DNU
 
 | |
6DNL
 
 | | Crystal Structure of Neisseria meningitidis DsbD c-terminal domain in the reduced form | | Descriptor: | ACETATE ION, Thiol:disulfide interchange protein DsbD, ZINC ION | | Authors: | Smith, R.P, Heras, B, Paxman, J.J. | | Deposit date: | 2018-06-06 | | Release date: | 2018-09-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and biochemical insights into the disulfide reductase mechanism of DsbD, an essential enzyme for neisserial pathogens.
J. Biol. Chem., 293, 2018
|
|
6JDS
 
 | |
