3AGF
 
 | |
3AGE
 
 | |
2DQX
 
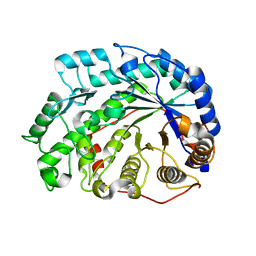 | | mutant beta-amylase (W55R) from soy bean | | Descriptor: | Beta-amylase | | Authors: | Ishikawa, K. | | Deposit date: | 2006-06-01 | | Release date: | 2007-05-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Kinetic and structural analysis of enzyme sliding on a substrate: multiple attack in beta-amylase
Biochemistry, 46, 2007
|
|
2CSX
 
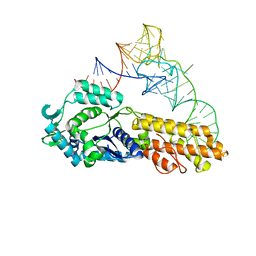 | | Crystal structure of Aquifex aeolicus methionyl-tRNA synthetase complexed with tRNA(Met) | | Descriptor: | Methionyl-tRNA synthetase, RNA (75-MER) | | Authors: | Nakanishi, K, Ogiso, Y, Nakama, T, Fukai, S, Nureki, O, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-23 | | Release date: | 2005-09-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for anticodon recognition by methionyl-tRNA synthetase.
Nat.Struct.Mol.Biol., 12, 2005
|
|
2CT8
 
 | | Crystal structure of Aquifex aeolicus methionyl-tRNA synthetase complexed with tRNA(Met) and methionyl-adenylate anologue | | Descriptor: | 5'-O-[(L-METHIONYL)-SULPHAMOYL]ADENOSINE, Methionyl-tRNA synthetase, RNA (74-MER) | | Authors: | Nakanishi, K, Ogiso, Y, Nakama, T, Fukai, S, Nureki, O, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-23 | | Release date: | 2005-09-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for anticodon recognition by methionyl-tRNA synthetase.
Nat.Struct.Mol.Biol., 12, 2005
|
|
3APV
 
 | | Crystal structure of the A variant of human alpha1-acid glycoprotein and amitriptyline complex | | Descriptor: | ACETIC ACID, Alpha-1-acid glycoprotein 2, Amitriptyline | | Authors: | Nishi, K, Ono, T, Nakamura, T, Fukunaga, N, Izumi, M, Watanabe, H, Suenaga, A, Maruyama, T, Yamagata, Y, Curry, S, Otagiri, M. | | Deposit date: | 2010-10-21 | | Release date: | 2011-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural insights into differences in drug-binding selectivity between two forms of human alpha1-acid glycoprotein genetic variants, the A and F1*S forms.
J. Biol. Chem., 286, 2011
|
|
3APX
 
 | | Crystal structure of the A variant of human alpha1-acid glycoprotein and chlorpromazine complex | | Descriptor: | 3-(2-chloro-10H-phenothiazin-10-yl)-N,N-dimethylpropan-1-amine, ACETIC ACID, Alpha-1-acid glycoprotein 2 | | Authors: | Nishi, K, Ono, T, Nakamura, T, Fukunaga, N, Izumi, M, Watanabe, H, Suenaga, A, Maruyama, T, Yamagata, Y, Curry, S, Otagiri, M. | | Deposit date: | 2010-10-21 | | Release date: | 2011-02-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into differences in drug-binding selectivity between two forms of human alpha1-acid glycoprotein genetic variants, the A and F1*S forms.
J. Biol. Chem., 286, 2011
|
|
3APW
 
 | | Crystal structure of the A variant of human alpha1-acid glycoprotein and disopyramide complex | | Descriptor: | Alpha-1-acid glycoprotein 2, Disopyramide | | Authors: | Nishi, K, Ono, T, Nakamura, T, Fukunaga, N, Izumi, M, Watanabe, H, Suenaga, A, Maruyama, T, Yamagata, Y, Curry, S, Otagiri, M. | | Deposit date: | 2010-10-21 | | Release date: | 2011-02-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into differences in drug-binding selectivity between two forms of human alpha1-acid glycoprotein genetic variants, the A and F1*S forms.
J. Biol. Chem., 286, 2011
|
|
3AK5
 
 | |
3APU
 
 | | Crystal structure of the A variant of human alpha1-acid glycoprotein | | Descriptor: | Alpha-1-acid glycoprotein 2, TETRAETHYLENE GLYCOL | | Authors: | Nishi, K, Ono, T, Nakamura, T, Fukunaga, N, Izumi, M, Watanabe, H, Suenaga, A, Maruyama, T, Yamagata, Y, Curry, S, Otagiri, M. | | Deposit date: | 2010-10-21 | | Release date: | 2011-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into differences in drug-binding selectivity between two forms of human alpha1-acid glycoprotein genetic variants, the A and F1*S forms.
J. Biol. Chem., 286, 2011
|
|
6U82
 
 | |
6CC2
 
 | | Crystal Structure of CDC45 from Entamoeba histolytica | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Cell division control protein 45 cdc45 putative, ... | | Authors: | Shi, K, Kurniawan, F, Kurahashi, K, Bielinsky, A, Aihara, H. | | Deposit date: | 2018-02-05 | | Release date: | 2018-06-27 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal Structure ofEntamoeba histolyticaCdc45 Suggests a Conformational Switch that May Regulate DNA Replication.
iScience, 3, 2018
|
|
6CA4
 
 | | Crystal structure of humanized D. rerio TDP2 by 14 mutations | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, MALONATE ION, ... | | Authors: | Shi, K, Aihrara, H. | | Deposit date: | 2018-01-29 | | Release date: | 2018-05-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.623 Å) | | Cite: | New fluorescence-based high-throughput screening assay for small molecule inhibitors of tyrosyl-DNA phosphodiesterase 2 (TDP2).
Eur J Pharm Sci, 118, 2018
|
|
6U81
 
 | | Crystal Structure of the Double Homeodomain of DUX4 in Complex with a DNA aptamer | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*CP*GP*TP*AP*AP*TP*CP*TP*AP*AP*TP*CP*AP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*TP*GP*AP*TP*TP*AP*GP*CP*CP*CP*AP*TP*TP*AP*CP*GP*C)-3'), ... | | Authors: | Shi, K, Aihara, H. | | Deposit date: | 2019-09-04 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | DNA aptamers against the DUX4 protein reveal novel therapeutic implications for FSHD.
Faseb J., 34, 2020
|
|
419D
 
 | |
7SPP
 
 | |
7SPO
 
 | | Crystal structure of the SARS-CoV-2 receptor binding domain in complex with VNAR 3B4 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, ... | | Authors: | Shi, K, Aihara, H. | | Deposit date: | 2021-11-02 | | Release date: | 2022-01-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Mechanisms of SARS-CoV-2 neutralization by shark variable new antigen receptors elucidated through X-ray crystallography.
Nat Commun, 12, 2021
|
|
5U0M
 
 | | Fatty aldehyde dehydrogenase from Marinobacter aquaeolei VT8 and cofactor complex | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, N-succinylglutamate 5-semialdehyde dehydrogenase, ... | | Authors: | Shi, K, Mulliner, K, Barney, B.M, Aihara, H. | | Deposit date: | 2016-11-24 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.075 Å) | | Cite: | Five Fatty Aldehyde Dehydrogenase Enzymes from Marinobacter and Acinetobacter spp. and Structural Insights into the Aldehyde Binding Pocket.
Appl. Environ. Microbiol., 83, 2017
|
|
5U0L
 
 | | X-ray crystal structure of fatty aldehyde dehydrogenase enzymes from Marinobacter aquaeolei VT8 complexed with a substrate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Shi, K, Mulliner, K, Barney, B.M, Aihara, H. | | Deposit date: | 2016-11-24 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.285 Å) | | Cite: | Five Fatty Aldehyde Dehydrogenase Enzymes from Marinobacter and Acinetobacter spp. and Structural Insights into the Aldehyde Binding Pocket.
Appl. Environ. Microbiol., 83, 2017
|
|
4F43
 
 | |
4F41
 
 | |
8V9Z
 
 | |
8V9Y
 
 | | X-ray crystal structure of nanobody JGFN4 | | Descriptor: | JGFN4 | | Authors: | Shi, K, Moller, N, Aihara, H. | | Deposit date: | 2023-12-10 | | Release date: | 2024-08-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Identification and biophysical characterization of a novel domain-swapped camelid antibody specific for fentanyl.
J.Biol.Chem., 300, 2024
|
|
8VA0
 
 | | X-ray crystal structure of JGFN4 N76D complexed with fentanyl in dimer form | | Descriptor: | 1,2-ETHANEDIOL, JGFN4, N-phenyl-N-[1-(2-phenylethyl)piperidin-4-yl]propanamide, ... | | Authors: | Shi, K, Moller, N, Aihara, H. | | Deposit date: | 2023-12-10 | | Release date: | 2024-08-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Identification and biophysical characterization of a novel domain-swapped camelid antibody specific for fentanyl.
J.Biol.Chem., 300, 2024
|
|
7TV2
 
 | | X-ray crystal structure of HIV-2 CA protein CTD | | Descriptor: | Capsid protein p24 | | Authors: | Shi, K, Aihara, H. | | Deposit date: | 2022-02-03 | | Release date: | 2023-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | HIV-2 Immature Particle Morphology Provides Insights into Gag Lattice Stability and Virus Maturation.
J.Mol.Biol., 435, 2023
|
|
