8JJ6
 
 | | Structure of the NELF-BCE complex | | Descriptor: | NELF-E, Negative elongation factor B, Negative elongation factor complex member C/D | | Authors: | Wang, Z, Cao, Y, Qin, Y. | | Deposit date: | 2023-05-29 | | Release date: | 2023-08-30 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structural basis of the human negative elongation factor NELF-B/C/E ternary complex.
Biochem.Biophys.Res.Commun., 677, 2023
|
|
4F1S
 
 | | Crystal structure of human PI3K-gamma in complex with a pyridyl-triazine-sulfonamide inhibitor | | Descriptor: | N-(5-{[3-(4-amino-6-methyl-1,3,5-triazin-2-yl)-5-(tetrahydro-2H-pyran-4-yl)pyridin-2-yl]amino}-2-chloropyridin-3-yl)methanesulfonamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Whittington, D.A, Tang, J, Yakowec, P. | | Deposit date: | 2012-05-07 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Synthesis and structure-activity relationships of dual PI3K/mTOR inhibitors based on a 4-amino-6-methyl-1,3,5-triazine sulfonamide scaffold.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
5LA7
 
 | | Crystal structure of human proheparanase, in complex with glucuronic acid configured aziridine probe JJB355 | | Descriptor: | (1~{S},2~{R},3~{S},4~{S},5~{S},6~{R})-2-(8-azidooctylamino)-3,4,5,6-tetrakis(oxidanyl)cyclohexane-1-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wu, L, Jin, Y, Davies, G.J. | | Deposit date: | 2016-06-13 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Activity-based probes for functional interrogation of retaining beta-glucuronidases.
Nat. Chem. Biol., 13, 2017
|
|
5L9Y
 
 | | Crystal structure of human heparanase, in complex with glucuronic acid configured aziridine probe JJB355 | | Descriptor: | (1~{S},2~{R},3~{S},4~{S},5~{S},6~{R})-2-(8-azidooctylamino)-3,4,5,6-tetrakis(oxidanyl)cyclohexane-1-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wu, L, Jin, Y, Davies, G.J. | | Deposit date: | 2016-06-13 | | Release date: | 2017-05-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Activity-based probes for functional interrogation of retaining beta-glucuronidases.
Nat. Chem. Biol., 13, 2017
|
|
4N39
 
 | |
3QK0
 
 | | Crystal structure of PI3K-gamma in complex with benzothiazole 82 | | Descriptor: | N-[6-(6-chloro-5-{[(4-fluorophenyl)sulfonyl]amino}pyridin-3-yl)-1,3-benzothiazol-2-yl]acetamide, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Whittington, D.A, Tang, J, Yakowec, P. | | Deposit date: | 2011-01-31 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Discovery and Optimization of a Series of Benzothiazole Phosphoinositide 3-Kinase (PI3K)/Mammalian Target of Rapamycin (mTOR) Dual Inhibitors.
J.Med.Chem., 54, 2011
|
|
4N3B
 
 | | Crystal Structure of human O-GlcNAc Transferase bound to a peptide from HCF-1 pro-repeat2(1-26)E10Q and UDP-5SGlcNAc | | Descriptor: | (2S,3R,4R,5S,6R)-3-(acetylamino)-4,5-dihydroxy-6-(hydroxymethyl)tetrahydro-2H-thiopyran-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate, Host cell factor 1, UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit | | Authors: | Lazarus, M.B, Herr, W, Walker, S. | | Deposit date: | 2013-10-06 | | Release date: | 2014-01-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | HCF-1 is cleaved in the active site of O-GlcNAc transferase.
Science, 342, 2013
|
|
4N3A
 
 | |
4N3C
 
 | |
4FLH
 
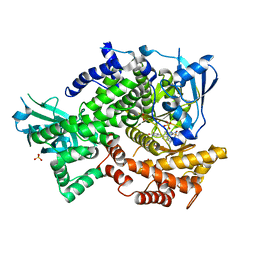 | | Crystal structure of human PI3K-gamma in complex with AMG511 | | Descriptor: | 4-(2-[(5-fluoro-6-methoxypyridin-3-yl)amino]-5-{(1R)-1-[4-(methylsulfonyl)piperazin-1-yl]ethyl}pyridin-3-yl)-6-methyl-1,3,5-triazin-2-amine, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Whittington, D.A, Tang, J, Yakowec, P. | | Deposit date: | 2012-06-14 | | Release date: | 2012-08-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Selective Class I Phosphoinositide 3-Kinase Inhibitors: Optimization of a Series of Pyridyltriazines Leading to the Identification of a Clinical Candidate, AMG 511.
J.Med.Chem., 55, 2012
|
|
9C5P
 
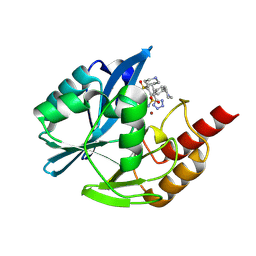 | | Inhibitor bound VIM1 | | Descriptor: | (2M)-4-(piperidine-4-sulfonyl)-4'-(piperidin-4-yl)-2-(1H-tetrazol-5-yl)[1,1'-biphenyl]-3-sulfonamide, MAGNESIUM ION, Metallo-beta-lactamase type 2, ... | | Authors: | Fischmann, T.O, Scapin, G. | | Deposit date: | 2024-06-06 | | Release date: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Discovery of sulfone containing metallo-beta-lactamase inhibitors with reduced bacterial cell efflux and histamine release issues
BMCL, 2024
|
|
8JB4
 
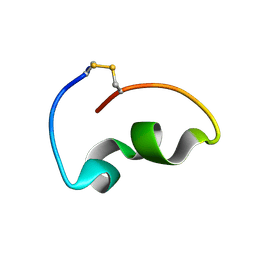 | | lipopolysaccharide-binding domain-LBDB | | Descriptor: | Antilipopolysaccharide factor D | | Authors: | Huang, J, Qin, Z. | | Deposit date: | 2023-05-08 | | Release date: | 2024-03-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Machine learning and genetic algorithm-guided directed evolution for the development of small-molecule antibiotics originating from antimicrobial peptides
To Be Published
|
|
8SDA
 
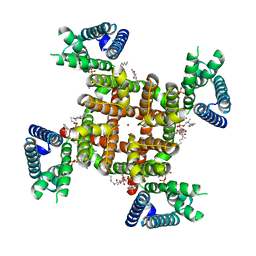 | | CryoEM structure of rat Kv2.1(1-598) L403A mutant in nanodiscs | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, POTASSIUM ION, Potassium voltage-gated channel subfamily B member 1 | | Authors: | Tan, X, Swartz, K.J. | | Deposit date: | 2023-04-06 | | Release date: | 2023-09-27 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Inactivation of the Kv2.1 channel through electromechanical coupling.
Nature, 622, 2023
|
|
8SD3
 
 | | CryoEM structure of rat Kv2.1(1-598) wild type in nanodiscs | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, POTASSIUM ION, Potassium voltage-gated channel subfamily B member 1 | | Authors: | Tan, X, Swartz, K.J. | | Deposit date: | 2023-04-06 | | Release date: | 2023-09-27 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Inactivation of the Kv2.1 channel through electromechanical coupling.
Nature, 622, 2023
|
|
6AF0
 
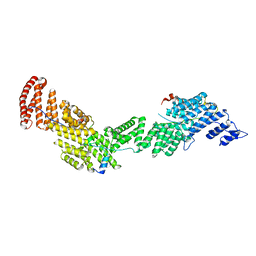 | | Structure of Ctr9, Paf1 and Cdc73 ternary complex from Myceliophthora thermophila | | Descriptor: | Cdc73 protein, Ctr9 protein, Paf1 protein | | Authors: | Wang, Z, Deng, P, Zhou, Y. | | Deposit date: | 2018-08-07 | | Release date: | 2018-09-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Transcriptional elongation factor Paf1 core complex adopts a spirally wrapped solenoidal topology.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6WUT
 
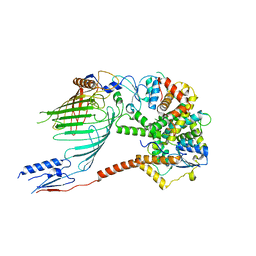 | |
6WUN
 
 | |
6WUL
 
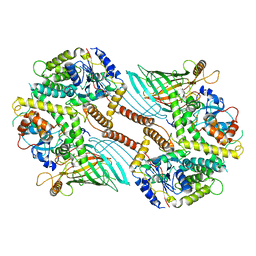 | | Mitochondrial SAM complex - dimer 1 in detergent | | Descriptor: | Bac_surface_Ag domain-containing protein, Sam35, Tom37 domain-containing protein | | Authors: | Ni, X, Botos, I, Diederichs, K. | | Deposit date: | 2020-05-04 | | Release date: | 2020-08-12 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insight into mitochondrial beta-barrel outer membrane protein biogenesis.
Nat Commun, 11, 2020
|
|
3T0T
 
 | | Crystal structure of S. aureus Pyruvate Kinase | | Descriptor: | N'-[(1E)-1-(1H-benzimidazol-2-yl)ethylidene]-5-bromo-2-hydroxybenzohydrazide, PHOSPHATE ION, Pyruvate kinase | | Authors: | Worrall, L.J, Vuckovic, M, Strynadka, N.C.J. | | Deposit date: | 2011-07-20 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Cheminformatics-driven discovery of selective, nanomolar inhibitors for staphylococcal pyruvate kinase.
Acs Chem.Biol., 7, 2012
|
|
4P6A
 
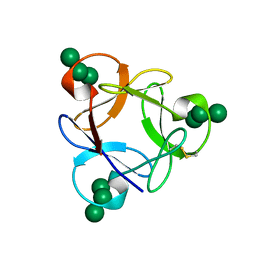 | | Crystal structure of a potent anti-HIV lectin actinohivin in complex with alpha-1,2-mannotriose | | Descriptor: | Actinohivin, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose | | Authors: | Zhang, F, Hoque, M.M, Suzuki, K, Tsunoda, M, Naomi, O, Tanaka, H, Takenaka, A. | | Deposit date: | 2014-03-23 | | Release date: | 2015-03-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.398 Å) | | Cite: | The characteristic structure of anti-HIV actinohivin in complex with three HMTG D1 chains of HIV-gp120.
Chembiochem, 15, 2014
|
|
6WUJ
 
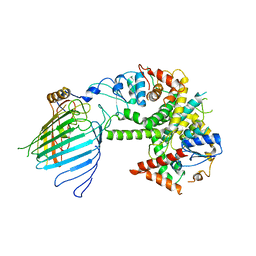 | | Mitochondrial SAM complex - monomer in detergent | | Descriptor: | Bac_surface_Ag domain-containing protein, Sam35, Tom37 domain-containing protein | | Authors: | Ni, X, Botos, I, Diederichs, K. | | Deposit date: | 2020-05-04 | | Release date: | 2020-08-12 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural insight into mitochondrial beta-barrel outer membrane protein biogenesis.
Nat Commun, 11, 2020
|
|
6WUH
 
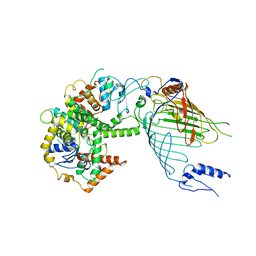 | | Mitochondrial SAM complex in lipid nanodiscs | | Descriptor: | Bac_surface_Ag domain-containing protein, Sam35, Tom37 domain-containing protein | | Authors: | Ni, X, Botos, I, Diederichs, K. | | Deposit date: | 2020-05-04 | | Release date: | 2020-08-12 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insight into mitochondrial beta-barrel outer membrane protein biogenesis.
Nat Commun, 11, 2020
|
|
6WUM
 
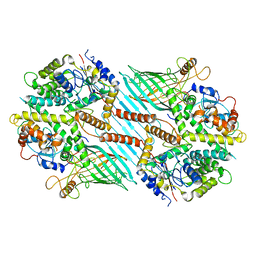 | | Mitochondrial SAM complex - dimer 2 in detergent | | Descriptor: | Bac_surface_Ag domain-containing protein, Sam35, Tom37 domain-containing protein | | Authors: | Ni, X, Botos, I, Diederichs, K. | | Deposit date: | 2020-05-04 | | Release date: | 2020-08-12 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insight into mitochondrial beta-barrel outer membrane protein biogenesis.
Nat Commun, 11, 2020
|
|
8IW4
 
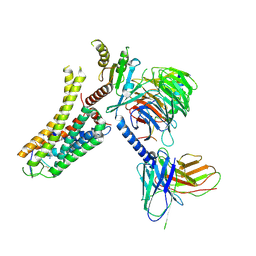 | | Cryo-EM structure of the SPE-bound mTAAR9-Gs complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Sun, J.P, Li, Q, Yang, F, Xu, Y.F, Guo, L.L, Lian, S, Zhang, M.H, Rong, N.K. | | Deposit date: | 2023-03-29 | | Release date: | 2023-05-31 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structural basis of amine odorant perception by a mammal olfactory receptor.
Nature, 618, 2023
|
|
8IW7
 
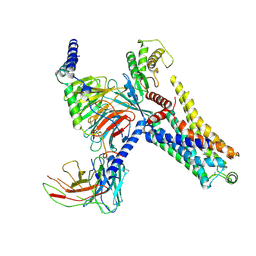 | | Cryo-EM structure of the PEA-bound mTAAR9-Gs complex | | Descriptor: | 2-PHENYLETHYLAMINE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Sun, J.P, Li, Q, Yang, F, Xu, Y.F, Guo, L.L, Lian, S, Zhang, M.H, Rong, N.K. | | Deposit date: | 2023-03-29 | | Release date: | 2023-05-31 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structural basis of amine odorant perception by a mammal olfactory receptor.
Nature, 618, 2023
|
|
