6BYS
 
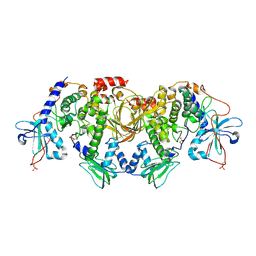 | | Structures of the PKA RI alpha holoenzyme with the FLHCC driver J-PKAc alpha or native PRKAc alpha | | Descriptor: | cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase type I-alpha regulatory subunit | | Authors: | Cao, B, Lu, T.W, Martinez Fiesco, J.A, Tomasini, M, Fan, L, Simon, S.M, Taylor, S.S, Zhang, P. | | Deposit date: | 2017-12-21 | | Release date: | 2019-04-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (4.75 Å) | | Cite: | Structures of the PKA RI alpha Holoenzyme with the FLHCC Driver J-PKAc alpha or Wild-Type PKAc alpha.
Structure, 27, 2019
|
|
8AOY
 
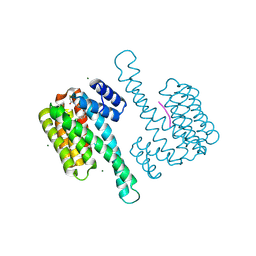 | | Small molecule stabilizer for ERalpha and 14-3-3 (1075478) | | Descriptor: | 14-3-3 protein sigma, 2-chloranyl-N-[3-[1-[4-[(4-chlorophenyl)amino]oxan-4-yl]carbonylpiperidin-4-yl]propyl]ethanamide, Estrogen receptor, ... | | Authors: | Konstantinidou, M, Visser, E.J, Vandenboorn, E.M.F, Sheng, C, Jaishankar, P, Overmans, M.J.A.M, Dutta, S, Neitz, J, Renslo, A, Ottmann, C, Brunsveld, L, Arkin, M. | | Deposit date: | 2022-08-09 | | Release date: | 2023-09-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-Based Optimization of Covalent, Small-Molecule Stabilizers of the 14-3-3 sigma /ER alpha Protein-Protein Interaction from Nonselective Fragments.
J.Am.Chem.Soc., 145, 2023
|
|
8AZE
 
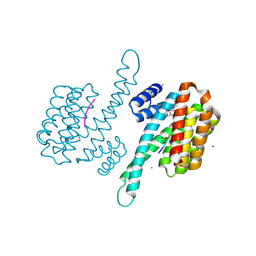 | | Small molecule stabilizer for ERalpha and 14-3-3 (1075306) | | Descriptor: | 14-3-3 protein sigma, 2-chloranyl-~{N}-[[1-[1-[(4-chlorophenyl)amino]cyclopentyl]carbonylpiperidin-4-yl]methyl]ethanamide, ERalpha peptide, ... | | Authors: | Konstantinidou, M, Visser, E.J, Vandenboorn, E.M.F, Sheng, C, Jaishankar, P, Overmans, M.J.A.M, Dutta, S, Neitz, J, Renslo, A, Ottmann, C, Brunsveld, L, Arkin, M. | | Deposit date: | 2022-09-06 | | Release date: | 2023-09-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Based Optimization of Covalent, Small-Molecule Stabilizers of the 14-3-3 sigma /ER alpha Protein-Protein Interaction from Nonselective Fragments.
J.Am.Chem.Soc., 145, 2023
|
|
8AXU
 
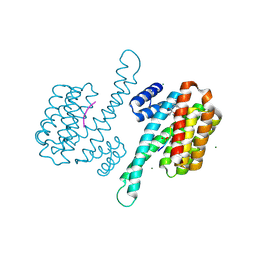 | | Small molecule stabilizer for ERalpha and 14-3-3 (1075297) | | Descriptor: | 14-3-3 protein sigma, 2-chloranyl-~{N}-[[1-[1-(4-chloranylphenoxy)cyclopentyl]carbonylpiperidin-4-yl]methyl]ethanamide, Estrogen receptor, ... | | Authors: | Konstantinidou, M, Visser, E.J, Vandenboorn, E.M.F, Sheng, C, Jaishankar, P, Overmans, M.J.A.M, Dutta, S, Neitz, J, Renslo, A, Ottmann, C, Brunsveld, L, Arkin, M. | | Deposit date: | 2022-09-01 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Based Optimization of Covalent, Small-Molecule Stabilizers of the 14-3-3 sigma /ER alpha Protein-Protein Interaction from Nonselective Fragments.
J.Am.Chem.Soc., 145, 2023
|
|
8AXE
 
 | | Small molecule stabilizer for ERalpha and 14-3-3 (1074210) | | Descriptor: | 14-3-3 protein sigma, 2-chloranyl-~{N}-[2-[1-[2-(4-chloranylphenoxy)-2-methyl-propanoyl]piperidin-4-yl]ethyl]ethanamide, Estrogen receptor, ... | | Authors: | Konstantinidou, M, Visser, E.J, Vandenboorn, E.M.F, Sheng, C, Jaishankar, P, Overmans, M.J.A.M, Dutta, S, Neitz, J, Renslo, A, Ottmann, C, Brunsveld, L, Arkin, M. | | Deposit date: | 2022-08-31 | | Release date: | 2023-09-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Based Optimization of Covalent, Small-Molecule Stabilizers of the 14-3-3 sigma /ER alpha Protein-Protein Interaction from Nonselective Fragments.
J.Am.Chem.Soc., 145, 2023
|
|
3J4F
 
 | | Structure of HIV-1 capsid protein by cryo-EM | | Descriptor: | capsid protein | | Authors: | Zhao, G, Perilla, J.R, Meng, X, Schulten, K, Zhang, P. | | Deposit date: | 2013-07-11 | | Release date: | 2013-07-24 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (8.6 Å) | | Cite: | Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics.
Nature, 497, 2013
|
|
3JA6
 
 | | Cryo-electron Tomography and All-atom Molecular Dynamics Simulations Reveal a Novel Kinase Conformational Switch in Bacterial Chemotaxis Signaling | | Descriptor: | Chemotaxis protein CheA, Chemotaxis protein CheW, Methyl-accepting chemotaxis protein 2 | | Authors: | Cassidy, C.K, Himes, B.A, Alvarez, F.J, Ma, J, Zhao, G, Perilla, J.R, Schulten, K, Zhang, P. | | Deposit date: | 2015-04-21 | | Release date: | 2015-12-09 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (12.7 Å) | | Cite: | CryoEM and computer simulations reveal a novel kinase conformational switch in bacterial chemotaxis signaling.
Elife, 4, 2015
|
|
6BYR
 
 | | Structures of the PKA RI alpha holoenzyme with the FLHCC driver J-PKAc alpha or native PKAc alpha | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DnaJ homolog subfamily B member 1,cAMP-dependent protein kinase catalytic subunit alpha chimera, MAGNESIUM ION, ... | | Authors: | Cao, B, Lu, T.W, Martinez Fiesco, J.A, Tomasini, M, Fan, L, Simon, S.M, Taylor, S.S, Zhang, P. | | Deposit date: | 2017-12-21 | | Release date: | 2019-04-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.661 Å) | | Cite: | Structures of the PKA RI alpha Holoenzyme with the FLHCC Driver J-PKAc alpha or Wild-Type PKAc alpha.
Structure, 27, 2019
|
|
3PSL
 
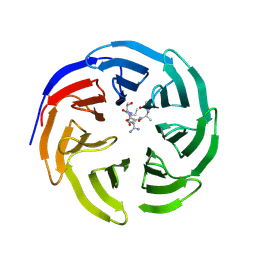 | | Fine-tuning the stimulation of MLL1 methyltransferase activity by a histone H3 based peptide mimetic | | Descriptor: | N-alpha acetylated form of histone H3, WD repeat-containing protein 5 | | Authors: | Avdic, V, Zhang, P, Lanouette, S, Voronova, A, Skerjanc, I, Couture, J.-F. | | Deposit date: | 2010-12-01 | | Release date: | 2010-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Fine-tuning the stimulation of MLL1 methyltransferase activity by a histone H3-based peptide mimetic.
Faseb J., 25, 2011
|
|
3J34
 
 | | Structure of HIV-1 Capsid Protein by Cryo-EM | | Descriptor: | capsid protein | | Authors: | Zhao, G, Perilla, J.R, Yufenyuy, E, Meng, X, Chen, B, Ning, J, Ahn, J, Gronenborn, A.M, Schulten, K, Aiken, C, Zhang, P. | | Deposit date: | 2013-02-23 | | Release date: | 2013-05-29 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (8.6 Å) | | Cite: | Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics.
Nature, 497, 2013
|
|
3J3Q
 
 | |
7LKI
 
 | |
3H4P
 
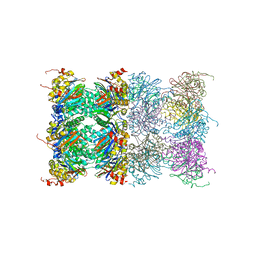 | | Proteasome 20S core particle from Methanocaldococcus jannaschii | | Descriptor: | Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Jeffrey, P.D, Zhang, F, Hu, M, Tian, G, Zhang, P, Finley, D, Shi, Y. | | Deposit date: | 2009-04-20 | | Release date: | 2009-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Structural Insights into the Regulatory Particle of the Proteasome from Methanocaldococcus jannaschii.
Mol.Cell, 34, 2009
|
|
4FSY
 
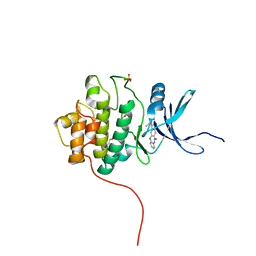 | | Crystal Structure of the CHK1 | | Descriptor: | 2-(11-oxo-10,11-dihydro-5H-dibenzo[b,e][1,4]diazepin-3-yl)benzamide, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Kang, Y.N, Stuckey, J.A, Chang, P, Russell, A.J. | | Deposit date: | 2012-06-27 | | Release date: | 2012-08-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the CHK1
To be Published
|
|
4FSM
 
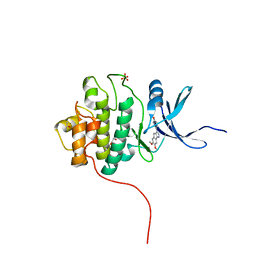 | | Crystal Structure of the CHK1 | | Descriptor: | 4-(6,7-dimethoxy-2,4-dihydroindeno[1,2-c]pyrazol-3-yl)phenol, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Kang, Y.N, Stuckey, J.A, Chang, P, Russell, A.J. | | Deposit date: | 2012-06-27 | | Release date: | 2012-08-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the CHK1
To be Published
|
|
4FT9
 
 | |
4FSZ
 
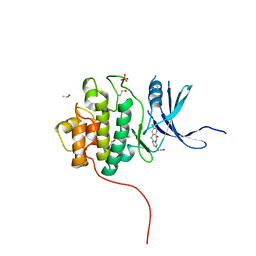 | | Crystal Structure of the CHK1 | | Descriptor: | (3-chloro-11-oxo-10,11-dihydro-5H-dibenzo[b,e][1,4]diazepin-8-yl)acetic acid, ISOPROPYL ALCOHOL, SULFATE ION, ... | | Authors: | Kang, Y.N, Stuckey, J.A, Chang, P, Russell, A.J. | | Deposit date: | 2012-06-27 | | Release date: | 2012-08-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the CHK1
To be Published
|
|
4FTA
 
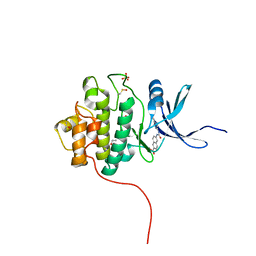 | | Crystal Structure of the CHK1 | | Descriptor: | 6-{[(5-cyanopyrazin-2-yl)carbamoyl]amino}naphthalene-2-carboxylic acid, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Kang, Y.N, Stuckey, J.A, Chang, P, Russell, A.J. | | Deposit date: | 2012-06-27 | | Release date: | 2012-08-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the CHK1
To be Published
|
|
4FTM
 
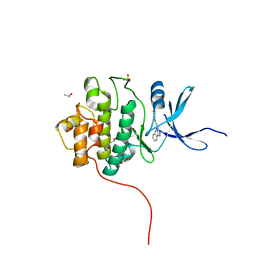 | | Crystal Structure of the CHK1 | | Descriptor: | 4-[2-(2,4-dihydroindeno[1,2-c]pyrazol-3-yl)ethyl]-2-methoxyphenol, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Kang, Y.N, Stuckey, J.A, Chang, P, Russell, A.J. | | Deposit date: | 2012-06-27 | | Release date: | 2012-08-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the CHK1
To be Published
|
|
4FTK
 
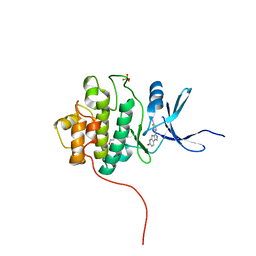 | | Crystal Structure of the CHK1 | | Descriptor: | 3-(4'-hydroxybiphenyl-4-yl)-2,4-dihydroindeno[1,2-c]pyrazol-6-ol, ISOPROPYL ALCOHOL, SULFATE ION, ... | | Authors: | Kang, Y.N, Stuckey, J.A, Chang, P, Russell, A.J. | | Deposit date: | 2012-06-27 | | Release date: | 2012-08-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the CHK1
To be Published
|
|
4FSN
 
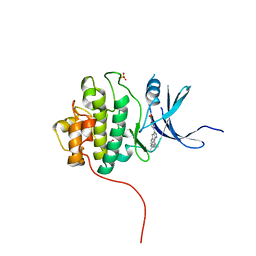 | | Crystal Structure of the CHK1 | | Descriptor: | 4-(6-{[(4-METHYLCYCLOHEXYL)AMINO]METHYL}-1,4-DIHYDROINDENO[1,2-C]PYRAZOL-3-YL)BENZOIC ACID, ISOPROPYL ALCOHOL, SULFATE ION, ... | | Authors: | Kang, Y.N, Stuckey, J.A, Chang, P, Russell, A.J. | | Deposit date: | 2012-06-27 | | Release date: | 2012-08-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the CHK1
To be Published
|
|
4FT3
 
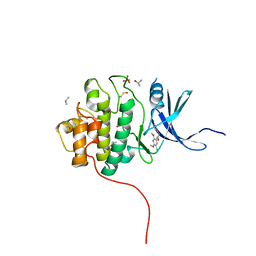 | | Crystal Structure of the CHK1 | | Descriptor: | 1-(5-chloro-2,4-dimethoxyphenyl)-3-pyrazin-2-ylurea, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Kang, Y.N, Stuckey, J.A, Chang, P, Russell, A.J. | | Deposit date: | 2012-06-27 | | Release date: | 2012-08-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the CHK1
To be Published
|
|
1KQ1
 
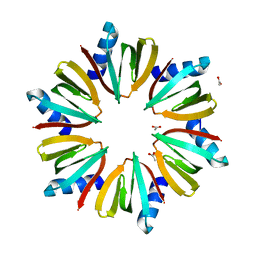 | | 1.55 A Crystal structure of the pleiotropic translational regulator, Hfq | | Descriptor: | ACETIC ACID, Host Factor for Q beta | | Authors: | Schumacher, M.A, Pearson, R.F, Moller, T, Valentin-Hansen, P, Brennan, R.G. | | Deposit date: | 2002-01-03 | | Release date: | 2002-07-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structures of the pleiotropic translational regulator Hfq and an Hfq-RNA complex: a bacterial Sm-like protein.
EMBO J., 21, 2002
|
|
1KQ2
 
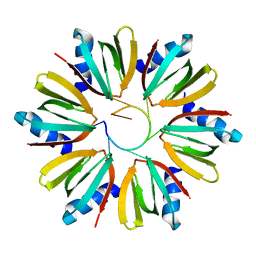 | | Crystal Structure of an Hfq-RNA Complex | | Descriptor: | 5'-R(*AP*UP*UP*UP*UP*UP*G)-3', Host factor for Q beta | | Authors: | Schumacher, M.A, Pearson, R.F, Moller, T, Valentin-Hansen, P, Brennan, R.G. | | Deposit date: | 2002-01-03 | | Release date: | 2002-07-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structures of the pleiotropic translational regulator Hfq and an Hfq-RNA complex: a bacterial Sm-like protein.
EMBO J., 21, 2002
|
|
7L7G
 
 | | Electron cryo-microscopy of the eukaryotic translation initiation factor 2B from Homo sapiens (updated model of PDB ID: 6CAJ) | | Descriptor: | 2-(4-chloranylphenoxy)-~{N}-[4-[2-(4-chloranylphenoxy)ethanoylamino]cyclohexyl]ethanamide, Translation initiation factor eIF-2B subunit alpha, Translation initiation factor eIF-2B subunit beta, ... | | Authors: | Tsai, J.C, Miller-Vedam, L.E, Anand, A, Jaishankar, P, Nguyen, H.C, Wang, L, Renslo, A.R, Frost, A, Walter, P. | | Deposit date: | 2020-12-28 | | Release date: | 2021-03-24 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | eIF2B conformation and assembly state regulates the integrated stress response.
Elife, 10, 2021
|
|
