8RKF
 
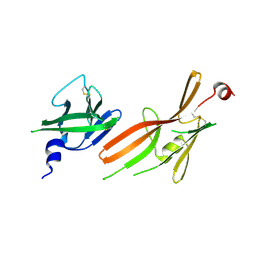 | | Crystal structure of the ZP-N1 and ZP-N2 domains of human ZP2 (hZP2-N1N2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Zona pellucida sperm-binding protein 2 | | Authors: | Dioguardi, E, Stsiapanava, A, de Sanctis, D, Jovine, L. | | Deposit date: | 2023-12-25 | | Release date: | 2024-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | ZP2 cleavage blocks polyspermy by modulating the architecture of the egg coat.
Cell, 187, 2024
|
|
4B3S
 
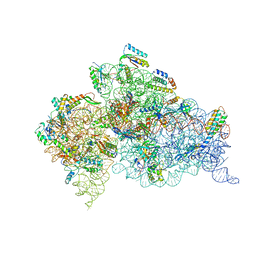 | | Crystal structure of the 30S ribosome in complex with compound 37 | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-diamino-2-{[3-O-(2,6-diamino-2,6-dideoxy-beta-L-idopyranosyl)-beta-D-ribofuranosyl]oxy}-3-hydroxycyclohexyl 2-amino-4-O-benzyl-2-deoxy-alpha-D-glucopyranoside, 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Ng, C.L, Lang, K, Shcherbakov, D, Matt, T, Perez-Fernandez, D, Patak, R, Meyer, M, Duscha, S, Akbergenov, R, Boukari, H, Freihofer, P, Kudyba, I, Reddy, M.S.K, Nandurikar, R.S, Ramakrishnan, V, Vasella, A, Bottger, E.C. | | Deposit date: | 2012-07-26 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | 4'-O-Substitutions Determine Selectivity of Aminoglycoside Antibiotics
Nat.Commun., 5, 2014
|
|
5NP8
 
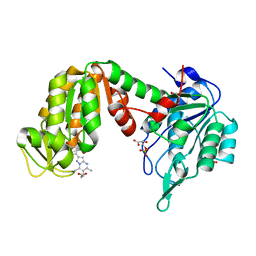 | | PGK1 in complex with CRT0063465 (3-[2-(4-bromophenyl)-5,7-dimethyl-pyrazolo[1,5-a]pyrimidin-6-yl]propanoic acid) | | Descriptor: | 1,2-ETHANEDIOL, 3-PHOSPHOGLYCERIC ACID, 3-[2-(4-bromophenyl)-5,7-dimethyl-pyrazolo[1,5-a]pyrimidin-6-yl]propanoic acid, ... | | Authors: | Turnbull, A.P, Bilsland, A.E, Liu, Y, Sumpton, D, Stevenson, K, Cairney, C.J, Roffey, J, Jenkinson, D, Keith, W.N. | | Deposit date: | 2017-04-13 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Novel Pyrazolopyrimidine Ligand of Human PGK1 and Stress Sensor DJ1 Modulates the Shelterin Complex and Telomere Length Regulation.
Neoplasia, 21, 2019
|
|
6HMB
 
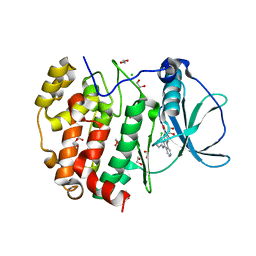 | | STRUCTURE OF PROTEIN KINASE CK2 CATALYTIC SUBUNIT (ISOFORM CK2ALPHA'; CSNK2A2 Gene product) IN COMPLEX WITH the inhibitor CX-4945 (Silmitasertib) | | Descriptor: | 1,2-ETHANEDIOL, 5-[(3-chlorophenyl)amino]benzo[c][2,6]naphthyridine-8-carboxylic acid, CHLORIDE ION, ... | | Authors: | Niefind, K, Lindenblatt, D, Applegate, V.M, Jose, J, Le Borgne, M. | | Deposit date: | 2018-09-12 | | Release date: | 2019-03-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Diacritic Binding of an Indenoindole Inhibitor by CK2 alpha Paralogs Explored by a Reliable Path to Atomic Resolution CK2 alpha ' Structures.
Acs Omega, 4, 2019
|
|
5LOQ
 
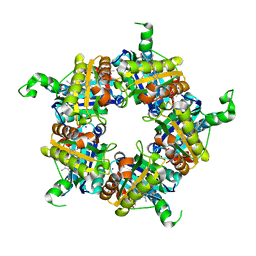 | | Structure of coproheme bound HemQ from Listeria monocytogenes | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,3,5,8-TETRAMETHYL-PORPHINE-2,4,6,7-TETRAPROPIONIC ACID FERROUS COMPLEX, Putative heme-dependent peroxidase lmo2113, ... | | Authors: | Puehringer, D, Mlynek, G, Hofbauer, S, Djinovic-Carugo, K, Obinger, C. | | Deposit date: | 2016-08-09 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Hydrogen peroxide-mediated conversion of coproheme to heme b by HemQ-lessons from the first crystal structure and kinetic studies.
FEBS J., 283, 2016
|
|
3DHA
 
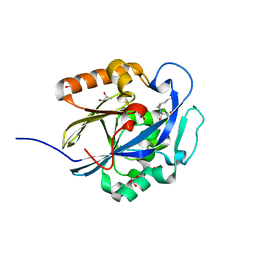 | | An Ultral High Resolution Structure of N-Acyl Homoserine Lactone Hydrolase with the Product N-Hexanoyl-L-Homoserine Bound at An Alternative Site | | Descriptor: | GLYCEROL, N-Acyl Homoserine Lactone Hydrolase, N-hexanoyl-L-homoserine, ... | | Authors: | Liu, D, Momb, J, Thomas, P.W, Moulin, A, Petsko, G.A, Fast, W, Ringe, D. | | Deposit date: | 2008-06-17 | | Release date: | 2008-07-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Mechanism of the quorum-quenching lactonase (AiiA) from Bacillus thuringiensis. 1. Product-bound structures.
Biochemistry, 47, 2008
|
|
8P1F
 
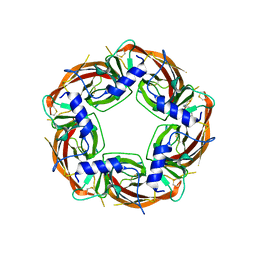 | | X-ray structure of acetylcholine-binding protein (AChBP) in complex with FL001909. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-azanyl-1-phenyl-piperidine-4-carboxylic acid, Acetylcholine-binding protein | | Authors: | Cederfelt, D, Boronat, P, Dobritzsch, D, Hennig, S, Fitzgerald, E.A, de Esch, I.J.P, Danielson, U.H. | | Deposit date: | 2023-05-12 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Elucidating the regulation of ligand gated ion channels via biophysical studies of ligand-induced conformational dynamics of acetylcholine binding proteins
To Be Published
|
|
8F5I
 
 | | SARS-CoV-2 S2 helix epitope scaffold bound by antibody DH1057.1 | | Descriptor: | DH1057.1 HC, DH1057.1 LC, Specialized acyl carrier protein, ... | | Authors: | Kapingidza, A.B, Wrapp, D, Winters, K, Azoitei, M.L. | | Deposit date: | 2022-11-14 | | Release date: | 2023-10-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Engineered immunogens to elicit antibodies against conserved coronavirus epitopes.
Nat Commun, 14, 2023
|
|
6WX1
 
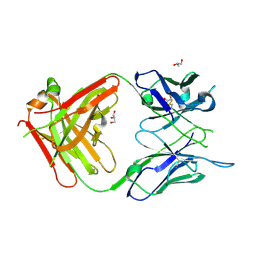 | | Antigen Binding Fragment of OKT9 | | Descriptor: | GLYCEROL, OKT9 Fab Heavy Chain, OKT9 Fab Light Chain | | Authors: | Rodriguez, J.A, Helguera, G, Sawaya, M, Cascio, D, Collazo, M, Flores, M, Zink, S, Ferrero, S, Payes, C, Fuentes, D, Short, C. | | Deposit date: | 2020-05-09 | | Release date: | 2021-05-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Antibody-Based Inhibition of Pathogenic New World Hemorrhagic Fever Mammarenaviruses by Steric Occlusion of the Human Transferrin Receptor 1 Apical Domain.
J.Virol., 95, 2021
|
|
8F5H
 
 | |
8F1F
 
 | | Structure of K48-linked tri-ubiquitin in complex with cyclic peptide | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, Non-proteinogenic cyclic peptide (inhibitor), ... | | Authors: | Lubkowski, J, Fushman, D, Lemma, B. | | Deposit date: | 2022-11-05 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mechanism of selective recognition of Lys48-linked polyubiquitin by macrocyclic peptide inhibitors of proteasomal degradation.
Nat Commun, 14, 2023
|
|
4A2S
 
 | | Structure of the engineered retro-aldolase RA95.5 | | Descriptor: | 1-(6-METHOXYNAPHTHALEN-2-YL)BUTANE-1,3-DIONE, INDOLE-3-GLYCEROL PHOSPHATE SYNTHASE | | Authors: | Giger, L, Caner, S, Kast, P, Baker, D, Ban, N, Hilvert, D. | | Deposit date: | 2011-09-28 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Evolution of a designed retro-aldolase leads to complete active site remodeling.
Nat.Chem.Biol., 9, 2013
|
|
4A12
 
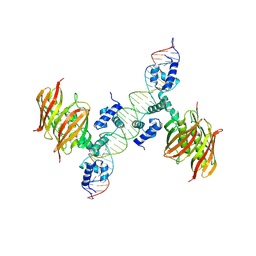 | | Structure of the global transcription regulator FapR from Staphylococcus aureus in complex with DNA operator | | Descriptor: | FAPR PROMOTER, TRANSCRIPTION FACTOR FAPR | | Authors: | Albanesi, D, Guerin, M.E, Buschiazzo, A, de Mendoza, D, Alzari, P.M. | | Deposit date: | 2011-09-13 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural Basis for Feed-Forward Transcriptional Regulation of Membrane Lipid Homeostasis in Staphylococcus Aureus.
Plos Pathog., 9, 2013
|
|
8P1E
 
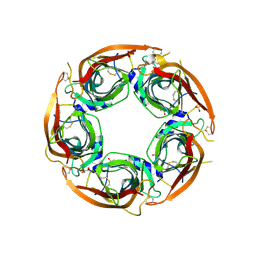 | | X-ray structure of acetylcholine-binding protein (AChBP) in complex with FL001613. | | Descriptor: | 1-[4-(trifluoromethyl)pyridin-2-yl]piperazine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholine-binding protein, ... | | Authors: | Cederfelt, D, Boronat, P, Dobritzsch, D, Hennig, S, Fitzgerald, E.A, de Esch, I.J.P, Danielson, U.H. | | Deposit date: | 2023-05-11 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Elucidating the regulation of ligand gated ion channels via biophysical studies of ligand-induced conformational dynamics of acetylcholine binding proteins
To Be Published
|
|
8ETN
 
 | |
2V54
 
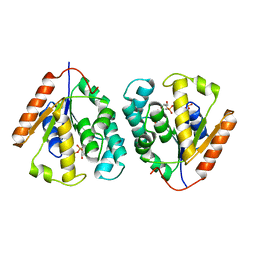 | | Crystal structure of vaccinia virus thymidylate kinase bound to TDP | | Descriptor: | MAGNESIUM ION, PYROPHOSPHATE 2-, THYMIDINE-5'-DIPHOSPHATE, ... | | Authors: | Caillat, C, Topalis, D, Agrofoglio, L.A, Pochet, S, Balzarini, J, Deville-Bonne, D, Meyer, P. | | Deposit date: | 2008-10-01 | | Release date: | 2008-10-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Poxvirus Thymidylate Kinase: An Unexpected Dimerization Has Implications for Antiviral Therapy
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2VLH
 
 | | Quinonoid intermediate of Citrobacter freundii tyrosine phenol-lyase formed with methionine | | Descriptor: | (2E)-2-{[(Z)-{3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4(1H)-YLIDENE}METHYL]IMINO}-4-(METHYLSULFANYL)BUTANOIC ACID, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, POTASSIUM ION, ... | | Authors: | Milic, D, Demidkina, T.V, Matkovic-Calogovic, D, Antson, A.A. | | Deposit date: | 2008-01-14 | | Release date: | 2008-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Insights Into the Catalytic Mechanism of Tyrosine Phenol-Lyase from X-Ray Structures of Quinonoid Intermediates.
J.Biol.Chem., 283, 2008
|
|
6DHP
 
 | | RT XFEL structure of the three-flash state of Photosystem II (3F, S0-rich) at 2.04 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Chatterjee, R, Young, I.D, Fuller, F.D, Lassalle, L, Ibrahim, M, Gul, S, Fransson, T, Brewster, A.S, Alonso-Mori, R, Hussein, R, Zhang, M, Douthit, L, de Lichtenberg, C, Cheah, M.H, Shevela, D, Wersig, J, Seufert, I, Sokaras, D, Pastor, E, Weninger, C, Kroll, T, Sierra, R.G, Aller, P, Butryn, A, Orville, A.M, Liang, M, Batyuk, A, Koglin, J.E, Carbajo, S, Boutet, S, Moriarty, N.W, Holton, J.M, Dobbek, H, Adams, P.D, Bergmann, U, Sauter, N.K, Zouni, A, Messinger, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2018-05-20 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structures of the intermediates of Kok's photosynthetic water oxidation clock.
Nature, 563, 2018
|
|
2VNF
 
 | | MOLECULAR BASIS OF HISTONE H3K4ME3 RECOGNITION BY ING4 | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, HISTONE H3, ... | | Authors: | Palacios, A, Munoz, I.G, Pantoja-Uceda, D, Marcaida, M.J, Torres, D, Martin-Garcia, J.M, Luque, I, Montoya, G, Blanco, F.J. | | Deposit date: | 2008-02-04 | | Release date: | 2008-04-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Molecular Basis of Histone H3K4Me3 Recognition by Ing4
J.Biol.Chem., 283, 2008
|
|
4A0X
 
 | | Structure of the global transcription regulator FapR from Staphylococcus aureus | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETATE ION, TRANSCRIPTION FACTOR FAPR, ... | | Authors: | Albanesi, D, Guerin, M.E, Buschiazzo, A, de Mendoza, D, Alzari, P.M. | | Deposit date: | 2011-09-13 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for Feed-Forward Transcriptional Regulation of Membrane Lipid Homeostasis in Staphylococcus Aureus.
Plos Pathog., 9, 2013
|
|
5FII
 
 | | Structure of a human aspartate kinase, chorismate mutase and TyrA domain. | | Descriptor: | PHENYLALANINE, PHENYLALANINE-4-HYDROXYLASE | | Authors: | Patel, D, Kopec, J, Shrestha, L, Fitzpatrick, F, Pinkas, D, Chaikuad, A, Dixon-Clarke, S, McCorvie, T.J, Burgess-Brown, N, von Delft, F, Arrowsmith, C, Edwards, A, Bountra, C, Yue, W.W. | | Deposit date: | 2015-09-25 | | Release date: | 2016-03-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Ligand-Dependent Dimerization of Phenylalanine Hydroxylase Regulatory Domain.
Sci.Rep., 6, 2016
|
|
4D1S
 
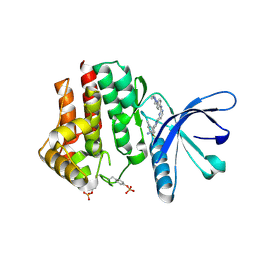 | | Pyrrole-3-carboxamides as potent and selective JAK2 inhibitors | | Descriptor: | 2-(5-chloro-2-methylphenyl)-1-methyl-5-(2-{[4-(4-methylpiperazin-1-yl)phenyl]amino}pyrimidin-4-yl)-1H-pyrrole-3-carboxamide, TYROSINE-PROTEIN KINASE JAK2 | | Authors: | Bertrand, J, Canevari, G, Fasolini, M, Brasca, M.G, Nesi, M, Avanzi, N, Ballinari, D, Bandiera, T, Bindi, S, Carenzi, D, Casero, D, Ceriani, L, Ciomei, M, Cirla, A, Colombo, M, Cribioli, S, Cristiani, C, Della Vedova, F, Fachin, G, Felder, E.R, Galvani, A, Isacchi, A, Mirizzi, D, Motto, I, Panzeri, A, Pesenti, E, Vianello, P, Gnocchi, P, Donati, D. | | Deposit date: | 2014-05-05 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Pyrrole-3-Carboxamides as Potent and Selective Jak2 Inhibitors.
Bioorg.Med.Chem., 22, 2014
|
|
8TXO
 
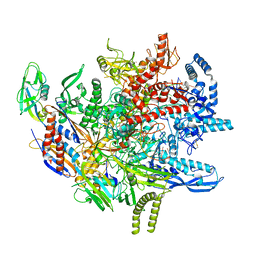 | | E. coli DNA-directed RNA polymerase transcription elongation complex bound to the unnatural dZ-PTP base pair in the active site | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Aiyer, S, Shan, Z, Oh, J, Wang, D, Tan, Y.Z, Lyumkis, D. | | Deposit date: | 2023-08-23 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Overcoming resolution attenuation during tilted cryo-EM data collection.
Nat Commun, 15, 2024
|
|
2VT7
 
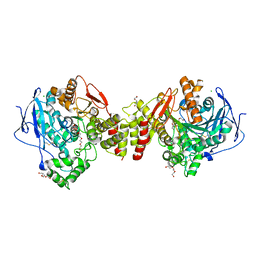 | | Native Torpedo californica acetylcholinesterase collected with a cumulated dose of 800000 Gy | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, CHLORIDE ION, ... | | Authors: | Colletier, J.P, Bourgeois, D, Sanson, B, Fournier, D, Sussman, J.L, Silman, I, Weik, M. | | Deposit date: | 2008-05-09 | | Release date: | 2008-07-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Shoot-and-Trap: Use of Specific X-Ray Damage to Study Structural Protein Dynamics by Temperature-Controlled Cryo-Crystallography.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
6C2Q
 
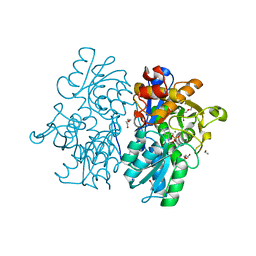 | | Crystal Structures of Cystathionine beta-Synthase from Saccharomyces cerevisiae: the Structure of the PLP-L-Serine Intermediate | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kreinbring, C.A, Tu, Y, Liu, D, Petsko, G.A, Ringe, D. | | Deposit date: | 2018-01-08 | | Release date: | 2018-04-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal Structures of Cystathionine beta-Synthase from Saccharomyces cerevisiae: One Enzymatic Step at a Time.
Biochemistry, 57, 2018
|
|
