8TYV
 
 | | Crystal structure of the SPX domain of XPR1 in complex with IP8 | | Descriptor: | (1R,3S,4R,5S,6R)-2,4,5,6-tetrakis(phosphonooxy)cyclohexane-1,3-diyl bis[trihydrogen (diphosphate)], Solute carrier family 53 member 1 | | Authors: | Wang, H, Shears, S.B. | | Deposit date: | 2023-08-25 | | Release date: | 2024-06-12 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Homeostatic coordination of cellular phosphate uptake and efflux requires an organelle-based receptor for the inositol pyrophosphate IP8.
Cell Rep, 43, 2024
|
|
8TYU
 
 | |
4QBS
 
 | |
4QBR
 
 | |
5EFW
 
 | | Crystal structure of LOV2-Zdk1 - the complex of oat LOV2 and the affibody protein Zdark1 | | Descriptor: | FLAVIN MONONUCLEOTIDE, NPH1-1, SULFATE ION, ... | | Authors: | Winkler, A, Wang, H, Hartmann, E, Hahn, K, Schlichting, I. | | Deposit date: | 2015-10-26 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | LOVTRAP: an optogenetic system for photoinduced protein dissociation.
Nat.Methods, 13, 2016
|
|
4I15
 
 | | Crystal structure of TbrPDEB1 | | Descriptor: | Class 1 phosphodiesterase PDEB1, MAGNESIUM ION, ZINC ION | | Authors: | Wang, H, Ke, H. | | Deposit date: | 2012-11-20 | | Release date: | 2013-03-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovery of Novel Trypanosoma brucei Phosphodiesterase B1 Inhibitors by Virtual Screening against the Unliganded TbrPDEB1 Crystal Structure.
J.Med.Chem., 56, 2013
|
|
5F7H
 
 | | Human T-cell immunoglobulin and mucin domain protein 4 (hTIM-4) complex with phosphoserine | | Descriptor: | CALCIUM ION, PHOSPHOSERINE, T-cell immunoglobulin and mucin domain-containing protein 4 | | Authors: | Gao, G.F, Lu, G, Wang, H, Qi, J. | | Deposit date: | 2015-12-08 | | Release date: | 2016-02-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of human TIM members: Ebolavirus entry-enhancing receptors
Chin.Sci.Bull., 60, 2015
|
|
8H7N
 
 | | Structure of nanobody 11A in complex with triazophos | | Descriptor: | 1,2-ETHANEDIOL, Nanobody 11A, SODIUM ION, ... | | Authors: | Wang, H, Li, J.D, Shen, X, Xu, Z.L, Sun, Y.M. | | Deposit date: | 2022-10-20 | | Release date: | 2023-10-25 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural Insights into the Stability and Recognition Mechanism of the Antiquinalphos Nanobody for the Detection of Quinalphos in Foods.
Anal.Chem., 95, 2023
|
|
8H7M
 
 | | Structure of nanobody 11A in complex with parathion | | Descriptor: | Nanobody 11A, parathion | | Authors: | Wang, H, Li, J.D, Shen, X, Xu, Z.L, Sun, Y.M. | | Deposit date: | 2022-10-20 | | Release date: | 2023-10-25 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural Insights into the Stability and Recognition Mechanism of the Antiquinalphos Nanobody for the Detection of Quinalphos in Foods.
Anal.Chem., 95, 2023
|
|
8H7I
 
 | | Structure of nanobody 11A in complex with quinalphos | | Descriptor: | Nanobody 11A, quinalphos | | Authors: | Wang, H, Li, J.D, Shen, X, Xu, Z.L, Sun, Y.M. | | Deposit date: | 2022-10-20 | | Release date: | 2023-10-25 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Insights into the Stability and Recognition Mechanism of the Antiquinalphos Nanobody for the Detection of Quinalphos in Foods.
Anal.Chem., 95, 2023
|
|
8H7R
 
 | | Structure of nanobody 11A in complex with coumaphos | | Descriptor: | Nanobody 11A, coumaphos | | Authors: | Wang, H, Li, J.D, Shen, X, Xu, Z.L, Sun, Y.M. | | Deposit date: | 2022-10-20 | | Release date: | 2023-10-25 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights into the Stability and Recognition Mechanism of the Antiquinalphos Nanobody for the Detection of Quinalphos in Foods.
Anal.Chem., 95, 2023
|
|
9BQJ
 
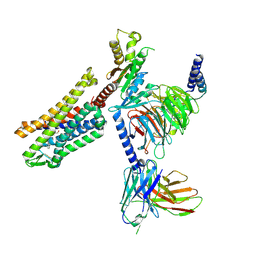 | | RO76 bound muOR-Gi1-scFv16 complex structure | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Wang, H, Majumdar, S, Kobilka, B.K. | | Deposit date: | 2024-05-10 | | Release date: | 2024-09-11 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Signaling Modulation Mediated by Ligand Water Interactions with the Sodium Site at mu OR.
Acs Cent.Sci., 10, 2024
|
|
8Y20
 
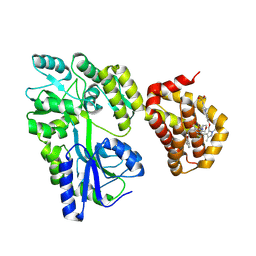 | | Crystal structure of the Mcl-1 in complex with A-1210477 | | Descriptor: | A-1210477, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Wang, H, Guo, M, Wei, H, Chen, Y. | | Deposit date: | 2024-01-25 | | Release date: | 2025-01-29 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Deciphering molecular specificity in MCL-1/BAK interaction and its implications for designing potent MCL-1 inhibitors.
Cell Death Differ., 32, 2025
|
|
8Y1Y
 
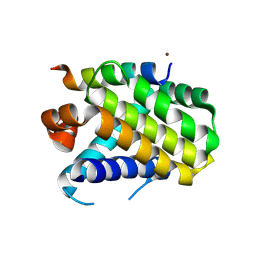 | | Crystal structure of the Mcl-1 in complex with a long BH3 peptide of BAK | | Descriptor: | BH3 peptide from Bcl-2 homologous antagonist/killer, Induced myeloid leukemia cell differentiation protein Mcl-1, ZINC ION | | Authors: | Wang, H, Guo, M, Wei, H, Chen, Y. | | Deposit date: | 2024-01-25 | | Release date: | 2025-01-29 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Deciphering molecular specificity in MCL-1/BAK interaction and its implications for designing potent MCL-1 inhibitors.
Cell Death Differ., 32, 2025
|
|
8Y1Z
 
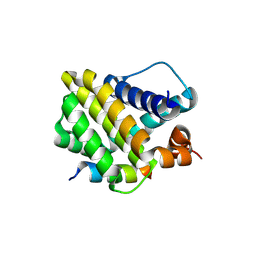 | | Crystal structure of the Mcl-1 in complex with a Short BH3 peptide of BAK | | Descriptor: | Induced myeloid leukemia cell differentiation protein Mcl-1, Short BH3 peptide from Bcl-2 homologous antagonist/killer | | Authors: | Wang, H, Guo, M, Wei, H, Chen, Y. | | Deposit date: | 2024-01-25 | | Release date: | 2025-01-29 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (1.914 Å) | | Cite: | Deciphering molecular specificity in MCL-1/BAK interaction and its implications for designing potent MCL-1 inhibitors.
Cell Death Differ., 32, 2025
|
|
4IQ8
 
 | | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase 3 from Saccharomyces cerevisiae | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase 3 | | Authors: | Wang, H, Liu, Q, Niu, L, Teng, M, Li, X. | | Deposit date: | 2013-01-11 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Preliminary crystallographic analysis of glyceraldehyde-3-phosphate dehydrogenase 3 from Saccharomyces cerevisiae.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
5F7F
 
 | |
5F71
 
 | | Human T-cell immunoglobulin and mucin domain protein 3 (hTIM-3) | | Descriptor: | Hepatitis A virus cellular receptor 2, SODIUM ION | | Authors: | Gao, G.F, Lu, G, Wang, H, Qi, J. | | Deposit date: | 2015-12-07 | | Release date: | 2016-02-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.404 Å) | | Cite: | Crystal structures of human TIM members: Ebolavirus entry-enhancing receptors
Chin.Sci.Bull., 60, 2015
|
|
5F70
 
 | | Crystal structures of human TIM members | | Descriptor: | Hepatitis A virus cellular receptor 1 | | Authors: | Gao, G.F, Lu, G, Wang, H, Qi, J. | | Deposit date: | 2015-12-07 | | Release date: | 2016-02-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of human TIM members: Ebolavirus entry-enhancing receptors
Chin.Sci.Bull., 60, 2015
|
|
8H3G
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) E166V Mutant in Complex with Inhibitor Enstrelvir | | Descriptor: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione, GLYCEROL | | Authors: | Wang, H, Lin, M, Duan, Y, Zhang, X, Zhou, H, Bian, Q, Liu, X, Rao, Z, Yang, H. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8H3L
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) Double Mutant (T21I and E166V) in Complex with Inhibitor Enstrelvir | | Descriptor: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | Authors: | Wang, H, Lin, M, Duan, Y, Zhang, X, Zhou, H, Bian, Q, Liu, X, Rao, Z, Yang, H. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8H3K
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) Double Mutant (L50F and E166V) in Complex with Inhibitor Enstrelvir | | Descriptor: | 3-(4-AMINO-2-METHYL-PYRIMIDIN-5-YLMETHYL)-5-(2-HYDROXY-ETHYL)-4-METHYL-THIAZOL-3-IUM, 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione, ... | | Authors: | Wang, H, Lin, M, Duan, Y, Zhang, X, Zhou, H, Bian, Q, Liu, X, Rao, Z, Yang, H. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
5WH5
 
 | |
5BYB
 
 | | Crystal structure of the catalytic domain of human diphosphoinositol pentakisphosphate kinase 2 (PPIP5K2) in complex with ADP and 1,5-(PA)2-IP4 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wang, H, Shears, S.B. | | Deposit date: | 2015-06-10 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Synthetic tools for studying the chemical biology of InsP8.
Chem.Commun.(Camb.), 51, 2015
|
|
5DGI
 
 | | Crystal structure of the catalytic domain of human diphosphoinositol pentakisphosphate kinase 2 (PPIP5K2) in complex with ADP and 3,5-(PCP)2-IP4 | | Descriptor: | 1,2-ETHANEDIOL, 3,5-di-methylenebisphosphonate inositol tetrakisphosphate, ACETATE ION, ... | | Authors: | Wang, H, Shears, S.B. | | Deposit date: | 2015-08-27 | | Release date: | 2016-08-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Cellular Cations Control Conformational Switching of Inositol Pyrophosphate Analogues.
Chemistry, 22, 2016
|
|
