1AXO
 
 | | STRUCTURAL ALIGNMENT OF THE (+)-TRANS-ANTI-[BP]DG ADDUCT POSITIONED OPPOSITE DC AT A DNA TEMPLATE-PRIMER JUNCTION, NMR, 6 STRUCTURES | | Descriptor: | 1,2,3-TRIHYDROXY-1,2,3,4-TETRAHYDROBENZO[A]PYRENE, DNA DUPLEX D(AAC-[BP]G-CTACCATCC)D(GGATGGTAGC) | | Authors: | Feng, B, Gorin, A.A, Hingerty, B.E, Geacintov, N.E, Broyde, S, Patel, D.J. | | Deposit date: | 1997-10-16 | | Release date: | 1998-07-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural alignment of the (+)-trans-anti-benzo[a]pyrene-dG adduct positioned opposite dC at a DNA template-primer junction.
Biochemistry, 36, 1997
|
|
6D9K
 
 | | Ternary RsAgo Complex with Guide RNA and Target DNA Containing A-G Non-canonical Pair | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, DNA (5'-D(P*TP*CP*GP*TP*CP*AP*CP*CP*TP*GP*GP*GP*CP*AP*GP*TP*AP*AP*C)-3'), ... | | Authors: | Liu, Y, Esyunina, D, Olovnikov, I, Teplova, M, Patel, D.J. | | Deposit date: | 2018-04-30 | | Release date: | 2018-07-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Accommodation of Helical Imperfections in Rhodobacter sphaeroides Argonaute Ternary Complexes with Guide RNA and Target DNA.
Cell Rep, 24, 2018
|
|
6D95
 
 | | Ternary RsAgo Complex with Guide RNA Paired and Target DNA containing A8-A8' Non-Canonical Pair | | Descriptor: | DNA 24-Mer, MAGNESIUM ION, RNA (5'-R(P*UP*UP*AP*CP*UP*GP*CP*AP*CP*AP*GP*GP*UP*GP*AP*CP*GP*A)-3'), ... | | Authors: | Liu, Y, Esyunina, D, Olovnikov, I, Teplova, M, Patel, D.J. | | Deposit date: | 2018-04-27 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Accommodation of Helical Imperfections in Rhodobacter sphaeroides Argonaute Ternary Complexes with Guide RNA and Target DNA.
Cell Rep, 24, 2018
|
|
1G70
 
 | | COMPLEX OF HIV-1 RRE-IIB RNA WITH RSG-1.2 PEPTIDE | | Descriptor: | HIV-1 RRE-IIB 32 NUCLEOTIDE RNA, RSG-1.2 PEPTIDE | | Authors: | Gosser, Y, Hermann, T, Majumdar, A, Hu, W, Frederick, R, Jiang, F, Xu, W, Patel, D.J. | | Deposit date: | 2000-11-08 | | Release date: | 2001-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Peptide-triggered conformational switch in HIV-1 RRE RNA complexes.
Nat.Struct.Biol., 8, 2001
|
|
1JVC
 
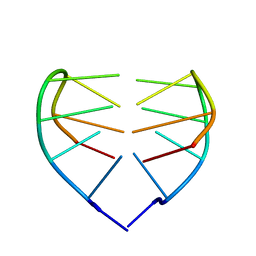 | | Dimeric DNA Quadruplex Containing Major Groove-Aligned A.T.A.T and G.C.G.C Tetrads Stabilized by Inter-Subunit Watson-Crick A:T and G:C Pairs | | Descriptor: | 5'-D(*GP*AP*GP*CP*AP*GP*GP*T)-3' | | Authors: | Zhang, N, Gorin, A, Majumdar, A, Kettani, A, Chernichenko, N, Skripkin, E, Patel, D.J. | | Deposit date: | 2001-08-29 | | Release date: | 2001-10-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Dimeric DNA quadruplex containing major groove-aligned A-T-A-T and G-C-G-C tetrads stabilized by inter-subunit Watson-Crick A-T and G-C pairs.
J.Mol.Biol., 312, 2001
|
|
1JJP
 
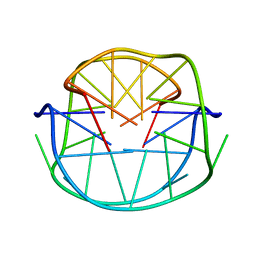 | | A(GGGG) Pentad-Containing Dimeric DNA Quadruplex Involving Stacked G(anti)G(anti)G(anti)G(syn) Tetrads | | Descriptor: | 5'-D(*GP*GP*GP*AP*GP*GP*TP*TP*TP*GP*GP*GP*AP*T)-3' | | Authors: | Zhang, N, Gorin, A, Majumdar, A, Kettani, A, Chernichenko, N, Skripkin, E, Patel, D.J. | | Deposit date: | 2001-07-09 | | Release date: | 2001-09-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | V-shaped scaffold: a new architectural motif identified in an A x (G x G x G x G) pentad-containing dimeric DNA quadruplex involving stacked G(anti) x G(anti) x G(anti) x G(syn) tetrads.
J.Mol.Biol., 311, 2001
|
|
1JO1
 
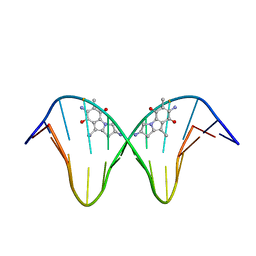 | | N7-Guanine Adduct of 2,7-diaminomitosene with DNA | | Descriptor: | 5'-D(*GP*TP*GP*(DAJ)GP*TP*AP*TP*AP*CP*CP*AP*C)-3', DECARBAMOYL-2,7-DIAMINOMITOSENE | | Authors: | Subramaniam, G, Paz, M.M, Kumar, G.S, Das, A, Palom, Y, Clement, C.C, Patel, D.J, Tomasz, M. | | Deposit date: | 2001-07-26 | | Release date: | 2001-09-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a guanine-N7-linked complex of the mitomycin C metabolite 2,7-diaminomitosene and DNA. Basis of sequence selectivity.
Biochemistry, 40, 2001
|
|
1B5K
 
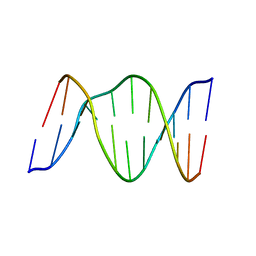 | | 3,N4-ETHENO-2'-DEOXYCYTIDINE OPPOSITE THYMIDINE IN AN 11-MER DUPLEX, SOLUTION STRUCTURE FROM NMR AND MOLECULAR DYNAMICS | | Descriptor: | DNA (5'-D(*CP*GP*TP*AP*CP*EDCP*CP*AP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*TP*GP*TP*GP*TP*AP*CP*G)-3') | | Authors: | Cullinan, D, Korobka, A, Grollman, A.P, Patel, D.J, Eisenberg, M, De Santos, C.L. | | Deposit date: | 1999-01-07 | | Release date: | 1999-01-13 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of an oligodeoxynucleotide duplex containing the exocyclic lesion 3,N4-etheno-2'-deoxycytidine opposite thymidine: comparison with the duplex containing deoxyadenosine opposite the adduct.
Biochemistry, 35, 1996
|
|
1AW4
 
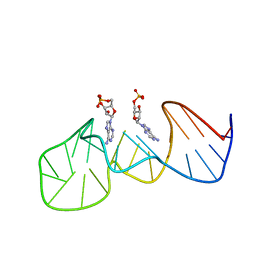 | |
1B6Y
 
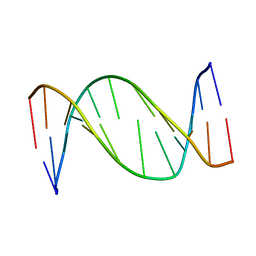 | | 3,N4-ETHENO-2'-DEOXYCYTIDINE OPPOSITE ADENINE IN AN 11-MER DUPLEX, SOLUTION STRUCTURE FROM NMR AND MOLECULAR DYNAMICS, 2 STRUCTURES | | Descriptor: | 5'-D(*CP*GP*TP*AP*CP*(EDC)P*CP*AP*TP*GP*C)-3', 5'-D(*GP*CP*AP*TP*GP*AP*GP*TP*AP*CP*G)-3' | | Authors: | Korobka, A, Cullinan, D, Cosman, M, Grollman, A.P, Patel, D.J, Eisenberg, M, De Los Santos, C. | | Deposit date: | 1999-01-19 | | Release date: | 1999-01-27 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an oligodeoxynucleotide duplex containing the exocyclic lesion 3,N4-etheno-2'-deoxycytidine opposite 2'-deoxyadenosine, determined by NMR spectroscopy and restrained molecular dynamics.
Biochemistry, 35, 1996
|
|
3U5O
 
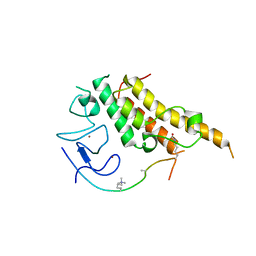 | |
3SUH
 
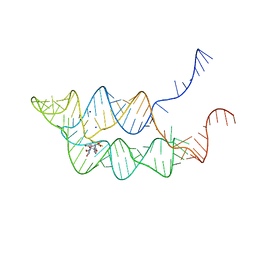 | | Crystal structure of THF riboswitch, bound with 5-formyl-THF | | Descriptor: | N-[4-({[(6S)-2-amino-5-formyl-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid, Riboswitch, SODIUM ION | | Authors: | Huang, L, Serganov, A, Patel, D.J. | | Deposit date: | 2011-07-11 | | Release date: | 2011-09-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Long-range pseudoknot interactions dictate the regulatory response in the tetrahydrofolate riboswitch.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3RAX
 
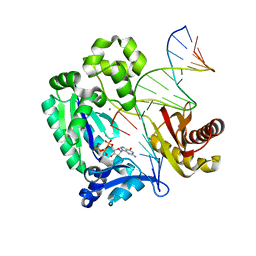 | |
3SLQ
 
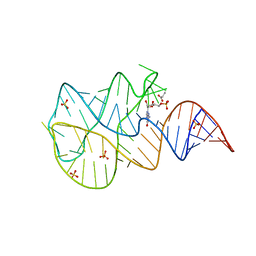 | | Crystal structure of the 2'- Deoxyguanosine riboswitch bound to guanosine-5'-monophosphate | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, RNA (68-MER), SUCCINIC ACID, ... | | Authors: | Pikovskaya, O, Polonskaia, A, Patel, D.J, Serganov, A. | | Deposit date: | 2011-06-24 | | Release date: | 2011-08-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural principles of nucleoside selectivity in a 2'-deoxyguanosine riboswitch.
Nat.Chem.Biol., 7, 2011
|
|
3SKR
 
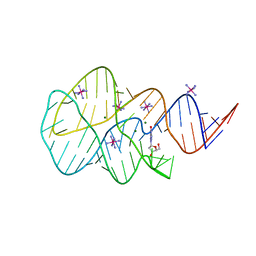 | | Crystal structure of the 2'- Deoxyguanosine riboswitch bound to 2'- Deoxyguanosine, cobalt Hexammine soak | | Descriptor: | 2'-DEOXY-GUANOSINE, COBALT HEXAMMINE(III), MAGNESIUM ION, ... | | Authors: | Pikovskaya, O, Polonskaia, A, Patel, D.J, Serganov, A. | | Deposit date: | 2011-06-23 | | Release date: | 2011-08-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural principles of nucleoside selectivity in a 2'-deoxyguanosine riboswitch.
Nat.Chem.Biol., 7, 2011
|
|
3SKZ
 
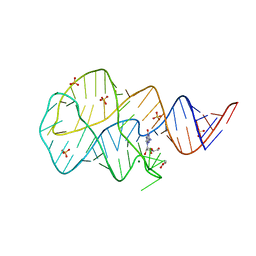 | | Crystal structure of the 2'- deoxyguanosine riboswitch bound to guanosine | | Descriptor: | GUANOSINE, MAGNESIUM ION, RNA (68-MER), ... | | Authors: | Pikovskaya, O, Polonskaia, A, Patel, D.J, Serganov, A. | | Deposit date: | 2011-06-23 | | Release date: | 2011-08-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.605 Å) | | Cite: | Structural principles of nucleoside selectivity in a 2'-deoxyguanosine riboswitch.
Nat.Chem.Biol., 7, 2011
|
|
3SOU
 
 | |
3RV0
 
 | | Crystal structure of K. polysporus Dcr1 without the C-terminal dsRBD | | Descriptor: | K. polysporus Dcr1, MAGNESIUM ION | | Authors: | Nakanishi, K, Weinberg, D.E, Bartel, D.P, Patel, D.J. | | Deposit date: | 2011-05-05 | | Release date: | 2011-08-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | The inside-out mechanism of dicers from budding yeasts.
Cell(Cambridge,Mass.), 146, 2011
|
|
3RZN
 
 | | Crystal Structure of Human Glycolipid Transfer Protein complexed with 3-O-sulfo-galactosylceramide containing nervonoyl acyl chain (24:1) | | Descriptor: | (15Z)-N-((1S,2R,3E)-2-HYDROXY-1-{[(3-O-SULFO-BETA-D-GALACTOPYRANOSYL)OXY]METHYL}HEPTADEC-3-ENYL)TETRACOS-15-ENAMIDE, Glycolipid transfer protein | | Authors: | Samygina, V, Cabo-Bilbao, A, Popov, A.N, Ochoa-Lizarralde, B, Patel, D.J, Brown, R.E, Malinina, L. | | Deposit date: | 2011-05-12 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Enhanced selectivity for sulfatide by engineered human glycolipid transfer protein.
Structure, 19, 2011
|
|
1D83
 
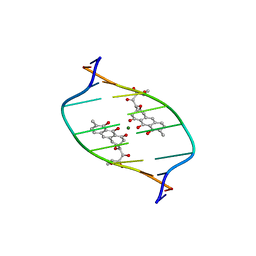 | | STRUCTURE REFINEMENT OF THE CHROMOMYCIN DIMER/DNA OLIGOMER COMPLEX IN SOLUTION | | Descriptor: | (1S)-5-deoxy-1-O-methyl-1-C-[(2R,3S)-3,5,7,10-tetrahydroxy-6-methyl-4-oxo-1,2,3,4-tetrahydroanthracen-2-yl]-D-xylulose, 2,6-dideoxy-4-O-methyl-alpha-D-galactopyranose-(1-3)-(2R,3R,6R)-6-hydroxy-2-methyltetrahydro-2H-pyran-3-yl acetate, 3-C-methyl-4-O-acetyl-alpha-L-Olivopyranose-(1-3)-(2R,5S,6R)-6-methyltetrahydro-2H-pyran-2,5-diol-(1-3)-(2R,5S,6R)-6-methyltetrahydro-2H-pyran-2,5-diol, ... | | Authors: | Gao, X, Mirau, P, Patel, D.J. | | Deposit date: | 1992-07-22 | | Release date: | 1993-07-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure refinement of the chromomycin dimer-DNA oligomer complex in solution.
J.Mol.Biol., 223, 1992
|
|
3SKL
 
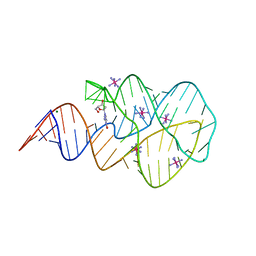 | | Crystal structure of the 2'- deoxyguanosine riboswitch bound to 2'-deoxyguanosine, iridium hexammine soak | | Descriptor: | 2'-DEOXY-GUANOSINE, IRIDIUM HEXAMMINE ION, MAGNESIUM ION, ... | | Authors: | Pikovskaya, O, Polonskaia, A, Patel, D.J, Serganov, A. | | Deposit date: | 2011-06-22 | | Release date: | 2011-08-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural principles of nucleoside selectivity in a 2'-deoxyguanosine riboswitch.
Nat.Chem.Biol., 7, 2011
|
|
3S0I
 
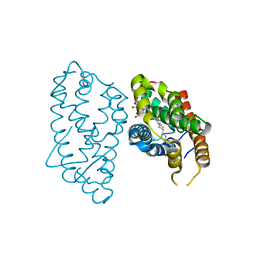 | | Crystal Structure of D48V mutant of Human Glycolipid Transfer Protein complexed with 3-O-sulfo galactosylceramide containing nervonoyl acyl chain | | Descriptor: | (15Z)-N-((1S,2R,3E)-2-HYDROXY-1-{[(3-O-SULFO-BETA-D-GALACTOPYRANOSYL)OXY]METHYL}HEPTADEC-3-ENYL)TETRACOS-15-ENAMIDE, Glycolipid transfer protein | | Authors: | Samygina, V, Popov, A.N, Cabo-Bilbao, A, Ochoa-Lizarralde, B, Patel, D.J, Brown, R.E, Malinina, L. | | Deposit date: | 2011-05-13 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Enhanced selectivity for sulfatide by engineered human glycolipid transfer protein.
Structure, 19, 2011
|
|
1D6D
 
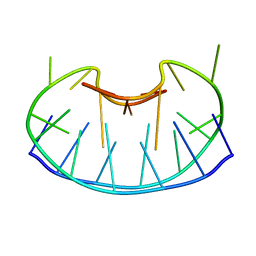 | | SOLUTION DNA STRUCTURE CONTAINING (A-A)-T TRIADS INTERDIGITATED BETWEEN A-T BASE PAIRS AND GGGG TETRADS; NMR, 8 STRUCT. | | Descriptor: | 5'-D(*AP*AP*GP*GP*TP*TP*TP*TP*AP*AP*GP*G)-3' | | Authors: | Kuryavyi, V.V, Kettani, A, Wang, W, Jones, R, Patel, D.J. | | Deposit date: | 1999-10-13 | | Release date: | 2000-01-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A diamond-shaped zipper-like DNA architecture containing triads sandwiched between mismatches and tetrads.
J.Mol.Biol., 295, 2000
|
|
3SOW
 
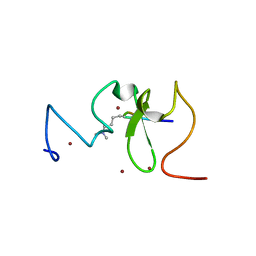 | |
3U5P
 
 | |
