473D
 
 | |
426D
 
 | |
2VA1
 
 | | Crystal structure of UMP kinase from Ureaplasma parvum | | Descriptor: | PHOSPHATE ION, URIDYLATE KINASE | | Authors: | Egeblad-Welin, L, Welin, M, Wang, L, Eriksson, S. | | Deposit date: | 2007-08-28 | | Release date: | 2007-09-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Functional Investigations of Ureaplasma Parvum Ump Kinase - a Potential Antibacterial Drug Target
FEBS J., 274, 2007
|
|
1A7C
 
 | | HUMAN PLASMINOGEN ACTIVATOR INHIBITOR TYPE-1 IN COMPLEX WITH A PENTAPEPTIDE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-D-ribopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, PENTAPEPTIDE, ... | | Authors: | Xue, Y, Inghardt, T, Sjolin, L, Deinum, J. | | Deposit date: | 1998-03-12 | | Release date: | 1999-03-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Interfering with the inhibitory mechanism of serpins: crystal structure of a complex formed between cleaved plasminogen activator inhibitor type 1 and a reactive-centre loop peptide
Structure, 6, 1998
|
|
1ENN
 
 | | SOLVENT ORGANIZATION IN AN OLIGONUCLEOTIDE CRYSTAL: THE STRUCTURE OF D(GCGAATTCG)2 AT ATOMIC RESOLUTION | | Descriptor: | CHLORIDE ION, DNA (5'-D(*GP*CP*GP*AP*AP*TP*TP*CP*G)-3'), MAGNESIUM ION, ... | | Authors: | Soler-Lopez, M, Malinina, L, Subirana, J.A. | | Deposit date: | 2000-03-21 | | Release date: | 2000-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Solvent organization in an oligonucleotide crystal. The structure of d(GCGAATTCG)2 at atomic resolution.
J.Biol.Chem., 275, 2000
|
|
1AZN
 
 | | CRYSTAL STRUCTURE OF THE AZURIN MUTANT PHE114ALA FROM PSEUDOMONAS AERUGINOSA AT 2.6 ANGSTROMS RESOLUTION | | Descriptor: | AZURIN, COPPER (II) ION | | Authors: | Tsai, L.-C, Sjolin, L, Langer, V, Pascher, T, Nar, H. | | Deposit date: | 1994-05-27 | | Release date: | 1994-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the azurin mutant Phe114Ala from Pseudomonas aeruginosa at 2.6 A resolution.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
1BY3
 
 | | FHUA FROM E. COLI | | Descriptor: | N-OCTYL-2-HYDROXYETHYL SULFOXIDE, PROTEIN (FERRICHROME-IRON RECEPTOR PRECURSOR (FHUA)) | | Authors: | Locher, K.P, Rees, B, Koebnik, R, Mitschler, A, Moulinier, L, Rosenbusch, J.P, Moras, D. | | Deposit date: | 1998-10-22 | | Release date: | 1999-01-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Transmembrane signaling across the ligand-gated FhuA receptor: crystal structures of free and ferrichrome-bound states reveal allosteric changes.
Cell(Cambridge,Mass.), 95, 1998
|
|
1C0A
 
 | | CRYSTAL STRUCTURE OF THE E. COLI ASPARTYL-TRNA SYNTHETASE : TRNAASP : ASPARTYL-ADENYLATE COMPLEX | | Descriptor: | ADENOSINE MONOPHOSPHATE, ASPARTYL TRNA, ASPARTYL TRNA SYNTHETASE, ... | | Authors: | Eiler, S, Dock-Bregeon, A.-C, Moulinier, L, Thierry, J.-C, Moras, D. | | Deposit date: | 1999-07-15 | | Release date: | 1999-11-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Synthesis of aspartyl-tRNA(Asp) in Escherichia coli--a snapshot of the second step.
EMBO J., 18, 1999
|
|
1B8A
 
 | | ASPARTYL-TRNA SYNTHETASE | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, PROTEIN (ASPARTYL-TRNA SYNTHETASE) | | Authors: | Schmitt, E, Moulinier, L, Thierry, J.-C, Moras, D. | | Deposit date: | 1999-01-27 | | Release date: | 1999-02-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of aspartyl-tRNA synthetase from Pyrococcus kodakaraensis KOD: archaeon specificity and catalytic mechanism of adenylate formation.
EMBO J., 17, 1998
|
|
1BY5
 
 | | FHUA FROM E. COLI, WITH ITS LIGAND FERRICHROME | | Descriptor: | FE (III) ION, FERRIC HYDROXAMATE UPTAKE PROTEIN, FERRICHROME, ... | | Authors: | Locher, K.P, Rees, B, Koebnik, R, Mitschler, A, Moulinier, L, Rosenbusch, J.P, Moras, D. | | Deposit date: | 1998-10-23 | | Release date: | 1999-01-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Transmembrane signaling across the ligand-gated FhuA receptor: crystal structures of free and ferrichrome-bound states reveal allosteric changes.
Cell(Cambridge,Mass.), 95, 1998
|
|
2TSA
 
 | | AZURIN MUTANT M121A | | Descriptor: | AZURIN, COPPER (II) ION | | Authors: | Tsai, L.-C, Bonander, N, Harata, K, Karlsson, B.G, Vanngard, T, Langer, V, Sjolin, L. | | Deposit date: | 1996-05-10 | | Release date: | 1996-11-08 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mutant Met121Ala of Pseudomonas aeruginosa azurin and its azide derivative: crystal structures and spectral properties.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
2TSB
 
 | | AZURIN MUTANT M121A-AZIDE | | Descriptor: | AZIDE ION, AZURIN AZIDE, COPPER (II) ION | | Authors: | Tsai, L.-C, Bonander, N, Harata, K, Karlsson, B.G, Vanngard, T, Langer, V, Sjolin, L. | | Deposit date: | 1996-05-10 | | Release date: | 1996-11-08 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mutant Met121Ala of Pseudomonas aeruginosa azurin and its azide derivative: crystal structures and spectral properties.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
2VNV
 
 | | Crystal structure of BclA lectin from burkholderia cenocepacia in complex with alpha-methyl-mannoside at 1.7 Angstrom resolution | | Descriptor: | BCLA, CALCIUM ION, SULFATE ION, ... | | Authors: | Lameignere, E, Malinovska, L, Mitchell, E.P, Imberty, A, Wimmerova, M. | | Deposit date: | 2008-02-07 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for Mannose Recognition by a Lectin from Opportunistic Bacteria Burkholderia Cenocepacia
Biochem.J., 411, 2008
|
|
6XCF
 
 | |
6XCB
 
 | |
6XCC
 
 | |
6XCD
 
 | |
6XCE
 
 | |
9EXW
 
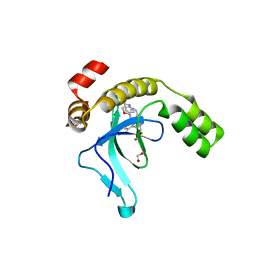 | | Crystal structure of the PWWP1 domain of NSD2 bound by compound 17. | | Descriptor: | 1,2-ETHANEDIOL, 7-[3-methyl-5-[2-methyl-5-[(pyridin-3-ylamino)methyl]phenyl]imidazol-4-yl]-4~{H}-1,4-benzoxazin-3-one, Histone-lysine N-methyltransferase NSD2 | | Authors: | Collie, G.W. | | Deposit date: | 2024-04-09 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Identification of Novel Potent NSD2-PWWP1 Ligands Using Structure-Based Design and Computational Approaches.
J.Med.Chem., 67, 2024
|
|
9EXX
 
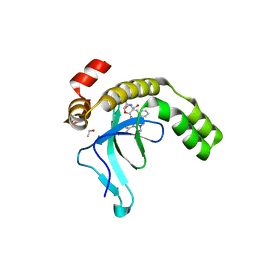 | | Crystal structure of the PWWP1 domain of NSD2 bound by compound 18. | | Descriptor: | 1,2-ETHANEDIOL, 4-methyl-3-[1-methyl-5-(3-oxidanylidene-4~{H}-1,4-benzoxazin-7-yl)imidazol-4-yl]-~{N}-phenyl-benzamide, ETHANOL, ... | | Authors: | Collie, G.W. | | Deposit date: | 2024-04-09 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.943 Å) | | Cite: | Identification of Novel Potent NSD2-PWWP1 Ligands Using Structure-Based Design and Computational Approaches.
J.Med.Chem., 67, 2024
|
|
9EXY
 
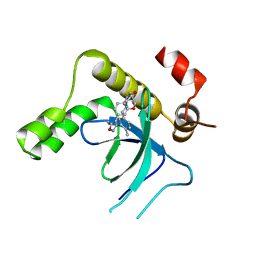 | | Crystal structure of the PWWP1 domain of NSD2 bound by compound 34. | | Descriptor: | 7-[5-methyl-3-[2-methyl-5-(piperidin-1-ylmethyl)phenyl]-1,2-oxazol-4-yl]-4~{H}-1,4-benzoxazin-3-one, Histone-lysine N-methyltransferase NSD2 | | Authors: | Collie, G.W. | | Deposit date: | 2024-04-09 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Identification of Novel Potent NSD2-PWWP1 Ligands Using Structure-Based Design and Computational Approaches.
J.Med.Chem., 67, 2024
|
|
3LAQ
 
 | | Structure-based engineering of species selectivity in the uPA-uPAR interaction | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Urokinase plasminogen activator surface receptor, Urokinase-type plasminogen activator | | Authors: | Huang, M. | | Deposit date: | 2010-01-06 | | Release date: | 2010-02-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure-based engineering of species selectivity in the interaction between urokinase and its receptor: implication for preclinical cancer therapy.
J.Biol.Chem., 285, 2010
|
|
2PQS
 
 | | Crystal Structure of the Bovine Lactadherin C2 Domain | | Descriptor: | Lactadherin | | Authors: | Huang, M, Furie, B.C. | | Deposit date: | 2007-05-02 | | Release date: | 2007-08-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the bovine lactadherin C2 domain, a membrane binding motif, shows similarity to the C2 domains of factor V and factor VIII.
J.Mol.Biol., 371, 2007
|
|
6TI5
 
 | | A New Structural Model of Abeta(1-40) Fibrils | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Bertini, I, Gonnelli, L, Luchinat, C, Mao, J, Nesi, A. | | Deposit date: | 2019-11-21 | | Release date: | 2020-07-22 | | Last modified: | 2024-06-19 | | Method: | SOLID-STATE NMR | | Cite: | Mixing A beta (1-40) and A beta (1-42) peptides generates unique amyloid fibrils.
Chem.Commun.(Camb.), 56, 2020
|
|
3P8X
 
 | | Synthesis, Structure, and Biological Activity of des-Side Chain Analogues of 1alpha,25-Dihydroxyvitamin D3 with Substituents at C-18 | | Descriptor: | (1R,3S,5Z)-5-{(2E)-2-[(3aR,7aS)-7a-(7-hydroxy-7-methyloctyl)octahydro-4H-inden-4-ylidene]ethylidene}-4-methylidenecyclohexane-1,3-diol, SULFATE ION, Vitamin D3 receptor | | Authors: | Rochel, N, Sato, Y, Moras, D. | | Deposit date: | 2010-10-15 | | Release date: | 2011-08-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Synthesis, Structure, and Biological Activity of des-Side Chain Analogues of 1 ,25-Dihydroxyvitamin D3 with Substituents at C-18
Chemmedchem, 6, 2011
|
|
